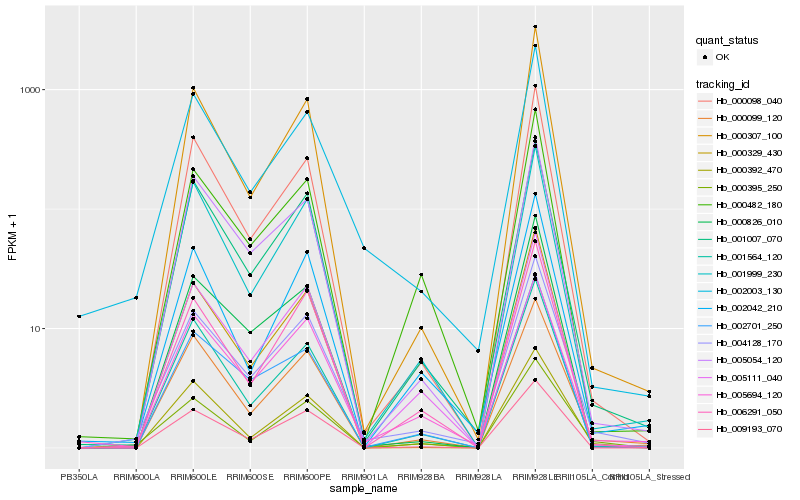
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004128_170 |
0.0 |
- |
- |
hypothetical protein POPTR_0016s08350g [Populus trichocarpa] |
| 2 |
Hb_001007_070 |
0.0789921304 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 3 |
Hb_002003_130 |
0.0854636016 |
- |
- |
hypothetical protein CISIN_1g027843mg [Citrus sinensis] |
| 4 |
Hb_000099_120 |
0.0857971724 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 5 |
Hb_005694_120 |
0.0871323324 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 6 |
Hb_000098_040 |
0.0887830841 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 7 |
Hb_005111_040 |
0.08929996 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 8 |
Hb_006291_050 |
0.0899924624 |
- |
- |
PREDICTED: polyribonucleotide nucleotidyltransferase 2, mitochondrial isoform X1 [Jatropha curcas] |
| 9 |
Hb_005054_120 |
0.0939756553 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000329_430 |
0.0940400065 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor UNE10 [Jatropha curcas] |
| 11 |
Hb_000307_100 |
0.0942428175 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_009193_070 |
0.0951776926 |
- |
- |
fatty acid elongase 3-ketoacyl-CoA synthase 1 [Arabidopsis thaliana] |
| 13 |
Hb_000826_010 |
0.0956794899 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001564_120 |
0.0973350702 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 15 |
Hb_002701_250 |
0.0973549629 |
- |
- |
PREDICTED: receptor-like protein kinase FERONIA [Jatropha curcas] |
| 16 |
Hb_000482_180 |
0.0978482972 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 17 |
Hb_001999_230 |
0.0978680812 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 18 |
Hb_002042_210 |
0.0981670632 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000395_250 |
0.0984227843 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 20 |
Hb_000392_470 |
0.1006328148 |
- |
- |
conserved hypothetical protein [Ricinus communis] |