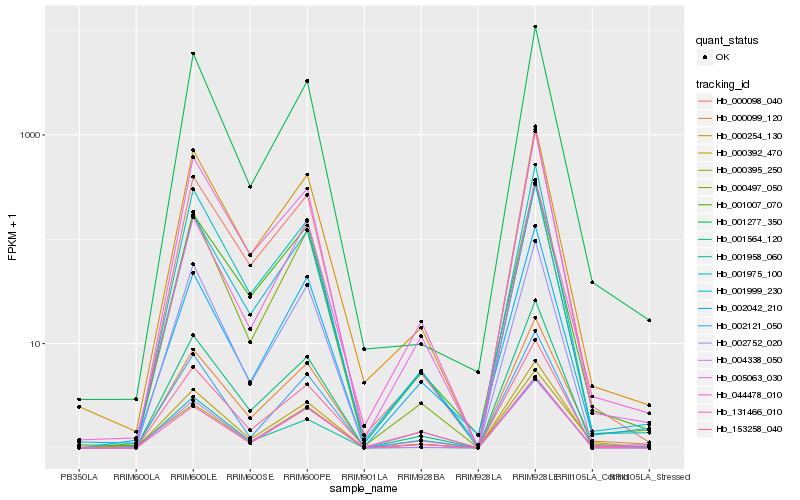
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000392_470 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000099_120 |
0.0518118514 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 3 |
Hb_001564_120 |
0.0624691724 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 4 |
Hb_004338_050 |
0.0683260331 |
- |
- |
hypothetical protein POPTR_0004s02760g [Populus trichocarpa] |
| 5 |
Hb_001999_230 |
0.0704346238 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 6 |
Hb_001007_070 |
0.0738566034 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 7 |
Hb_000497_050 |
0.0778567075 |
- |
- |
PREDICTED: photosystem II repair protein PSB27-H1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_005063_030 |
0.0789059812 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000098_040 |
0.0792350183 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000254_130 |
0.0818888613 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_131466_010 |
0.0822471814 |
- |
- |
PREDICTED: putative amidase C869.01 [Jatropha curcas] |
| 12 |
Hb_001975_100 |
0.0828380956 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000395_250 |
0.083480965 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 12 [Jatropha curcas] |
| 14 |
Hb_001277_350 |
0.0844869278 |
- |
- |
early light-induced protein, putative [Ricinus communis] |
| 15 |
Hb_153258_040 |
0.0862484897 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 16 |
Hb_044478_010 |
0.086636428 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 17 |
Hb_002121_050 |
0.0871631276 |
- |
- |
hypothetical protein EUGRSUZ_D02586 [Eucalyptus grandis] |
| 18 |
Hb_001958_060 |
0.0886055863 |
- |
- |
PREDICTED: calcium uptake protein 1, mitochondrial-like [Jatropha curcas] |
| 19 |
Hb_002042_210 |
0.0887276967 |
- |
- |
PREDICTED: UPF0603 protein At1g54780, chloroplastic [Jatropha curcas] |
| 20 |
Hb_002752_020 |
0.0902080473 |
- |
- |
FKBP-type peptidyl-prolyl cis-trans isomerase 3 family protein [Populus trichocarpa] |