| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
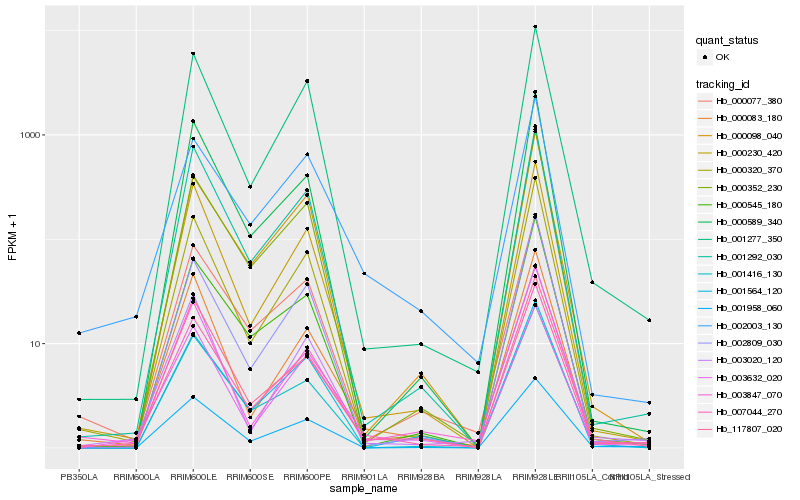
Hb_007044_270 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000320_370 |
0.0686791463 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002809_030 |
0.0759867292 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_001958_060 |
0.0784323114 |
- |
- |
PREDICTED: calcium uptake protein 1, mitochondrial-like [Jatropha curcas] |
| 5 |
Hb_117807_020 |
0.0804969434 |
- |
- |
methyltransferase, putative [Ricinus communis] |
| 6 |
Hb_000230_420 |
0.0805285906 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 7 |
Hb_001277_350 |
0.0805821887 |
- |
- |
early light-induced protein, putative [Ricinus communis] |
| 8 |
Hb_003020_120 |
0.081916891 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 9 |
Hb_000077_380 |
0.0819670759 |
- |
- |
PREDICTED: protochlorophyllide reductase [Jatropha curcas] |
| 10 |
Hb_000352_230 |
0.0833283698 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 11 |
Hb_000545_180 |
0.083680484 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 12 |
Hb_000083_180 |
0.0847794194 |
- |
- |
PREDICTED: probable 2-oxoglutarate/Fe(II)-dependent dioxygenase [Jatropha curcas] |
| 13 |
Hb_000589_340 |
0.0848699621 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 14 |
Hb_003632_020 |
0.0864280322 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 15 |
Hb_001292_030 |
0.0864793522 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003847_070 |
0.0865604251 |
- |
- |
PREDICTED: violaxanthin de-epoxidase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001564_120 |
0.0872461107 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 18 |
Hb_002003_130 |
0.0885017345 |
- |
- |
hypothetical protein CISIN_1g027843mg [Citrus sinensis] |
| 19 |
Hb_000098_040 |
0.0888504858 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 20 |
Hb_001416_130 |
0.0888801953 |
- |
- |
conserved hypothetical protein [Ricinus communis] |