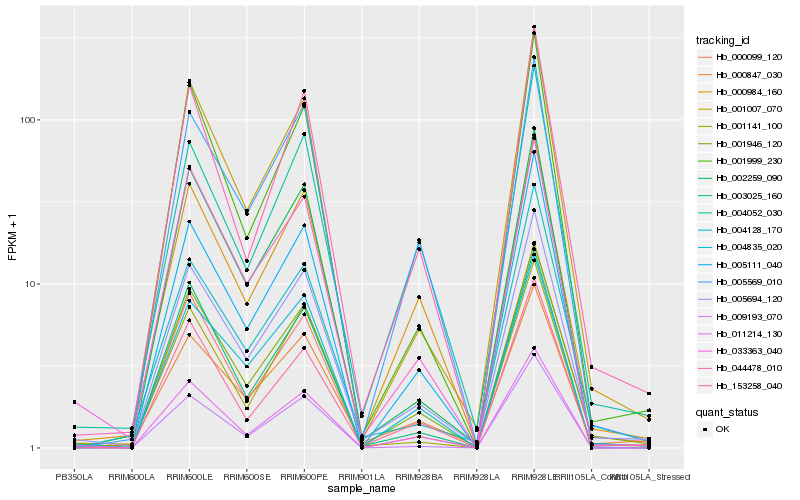
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005694_120 |
0.0 |
- |
- |
PREDICTED: calcium-transporting ATPase, endoplasmic reticulum-type [Jatropha curcas] |
| 2 |
Hb_001946_120 |
0.0434200898 |
- |
- |
PREDICTED: SPX domain-containing protein 1-like [Jatropha curcas] |
| 3 |
Hb_000847_030 |
0.0557929248 |
- |
- |
PREDICTED: asparagine--tRNA ligase, cytoplasmic 2 [Jatropha curcas] |
| 4 |
Hb_001007_070 |
0.0685422377 |
- |
- |
thioredoxin m(mitochondrial)-type, putative [Ricinus communis] |
| 5 |
Hb_005111_040 |
0.0747492917 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 6 |
Hb_044478_010 |
0.078306901 |
- |
- |
plastoquinol-plastocyanin reductase, putative [Ricinus communis] |
| 7 |
Hb_000984_160 |
0.0811426286 |
- |
- |
PREDICTED: amino acid permease 6-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_153258_040 |
0.0832412514 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 9 |
Hb_001999_230 |
0.0839160818 |
- |
- |
PREDICTED: uncharacterized protein LOC105630678 [Jatropha curcas] |
| 10 |
Hb_002259_090 |
0.0843931335 |
- |
- |
PREDICTED: ABC transporter G family member 11 [Jatropha curcas] |
| 11 |
Hb_005569_010 |
0.0847673772 |
- |
- |
Heme-binding protein, putative [Ricinus communis] |
| 12 |
Hb_004835_020 |
0.085740968 |
- |
- |
PREDICTED: receptor-like protein 12 [Populus euphratica] |
| 13 |
Hb_011214_130 |
0.0865772766 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 86 [Jatropha curcas] |
| 14 |
Hb_004128_170 |
0.0871323324 |
- |
- |
hypothetical protein POPTR_0016s08350g [Populus trichocarpa] |
| 15 |
Hb_033363_040 |
0.0881940089 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |
| 16 |
Hb_000099_120 |
0.0886688022 |
- |
- |
PREDICTED: GDT1-like protein 1, chloroplastic isoform X3 [Jatropha curcas] |
| 17 |
Hb_004052_030 |
0.0920056834 |
- |
- |
PREDICTED: uncharacterized protein LOC105640637 [Jatropha curcas] |
| 18 |
Hb_003025_160 |
0.0922962255 |
- |
- |
Protein HVA22, putative [Ricinus communis] |
| 19 |
Hb_001141_100 |
0.094077479 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2-like [Jatropha curcas] |
| 20 |
Hb_009193_070 |
0.0951506685 |
- |
- |
fatty acid elongase 3-ketoacyl-CoA synthase 1 [Arabidopsis thaliana] |