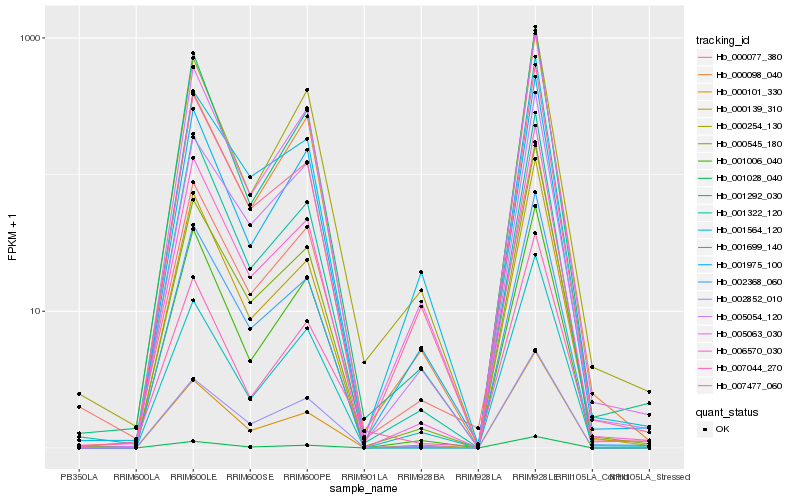
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000077_380 |
0.0 |
- |
- |
PREDICTED: protochlorophyllide reductase [Jatropha curcas] |
| 2 |
Hb_005063_030 |
0.0573844176 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 3 |
Hb_000101_330 |
0.064268421 |
- |
- |
- |
| 4 |
Hb_006570_030 |
0.064787431 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 5 |
Hb_001975_100 |
0.0651476736 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 6 |
Hb_001564_120 |
0.0688758731 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 7 |
Hb_002852_010 |
0.0702932475 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 8 |
Hb_005054_120 |
0.0739496942 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000254_130 |
0.0740761985 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 10 |
Hb_000545_180 |
0.0741940748 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 11 |
Hb_007477_060 |
0.0742281184 |
- |
- |
unknown [Medicago truncatula] |
| 12 |
Hb_002368_060 |
0.0746154241 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_001028_040 |
0.0748460488 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |
| 14 |
Hb_001292_030 |
0.0753561676 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 15 |
Hb_007044_270 |
0.0819670759 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 16 |
Hb_000098_040 |
0.0824636724 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001322_120 |
0.0830955223 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |
| 18 |
Hb_001699_140 |
0.0834172428 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000139_310 |
0.083590181 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 20 |
Hb_001006_040 |
0.0850116709 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |