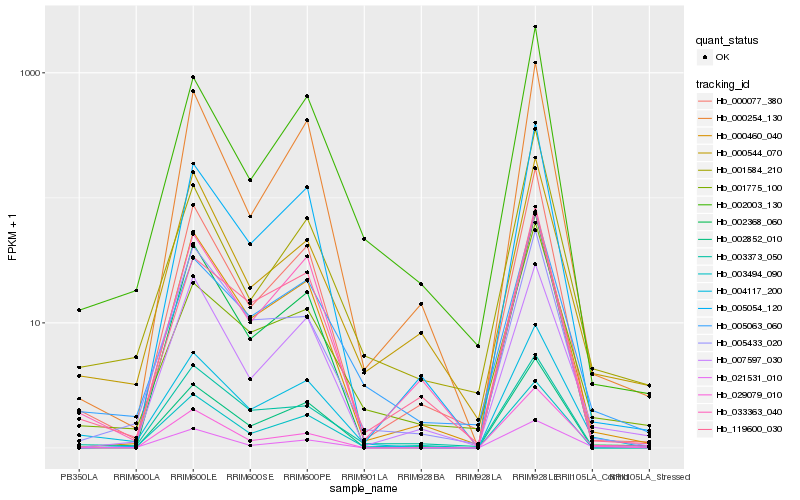
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004117_200 |
0.0 |
- |
- |
PREDICTED: RING-H2 finger protein ATL74 [Jatropha curcas] |
| 2 |
Hb_005063_060 |
0.1095371425 |
transcription factor |
TF Family: GNAT |
PREDICTED: uncharacterized protein LOC105647704 [Jatropha curcas] |
| 3 |
Hb_002852_010 |
0.1269864671 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 4 |
Hb_021531_010 |
0.1278110892 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 5 |
Hb_000077_380 |
0.1304284745 |
- |
- |
PREDICTED: protochlorophyllide reductase [Jatropha curcas] |
| 6 |
Hb_000460_040 |
0.133476267 |
- |
- |
PREDICTED: protein phosphatase 2C 57 [Jatropha curcas] |
| 7 |
Hb_002003_130 |
0.1388951023 |
- |
- |
hypothetical protein CISIN_1g027843mg [Citrus sinensis] |
| 8 |
Hb_003373_050 |
0.1399101258 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |
| 9 |
Hb_005433_020 |
0.1403219007 |
- |
- |
PREDICTED: multicopper oxidase LPR1 [Jatropha curcas] |
| 10 |
Hb_119600_030 |
0.1406490134 |
- |
- |
PREDICTED: uncharacterized protein LOC105635187 [Jatropha curcas] |
| 11 |
Hb_003494_090 |
0.1433409008 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 12 |
Hb_005054_120 |
0.1435170724 |
- |
- |
PREDICTED: sedoheptulose-1,7-bisphosphatase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_002368_060 |
0.1440401555 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_001775_100 |
0.1442893922 |
- |
- |
PREDICTED: probable 2-carboxy-D-arabinitol-1-phosphatase [Jatropha curcas] |
| 15 |
Hb_001584_210 |
0.145970671 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 16 |
Hb_007597_030 |
0.1459744432 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 17 |
Hb_000254_130 |
0.1467965537 |
- |
- |
Photosystem I reaction center subunit VI, chloroplast precursor, putative [Ricinus communis] |
| 18 |
Hb_000544_070 |
0.1468467794 |
- |
- |
Peptidyl-prolyl cis-trans isomerase FKBP17-2 [Morus notabilis] |
| 19 |
Hb_029079_010 |
0.1474449077 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 20 |
Hb_033363_040 |
0.1477255541 |
- |
- |
PREDICTED: probable galacturonosyltransferase-like 3 [Jatropha curcas] |