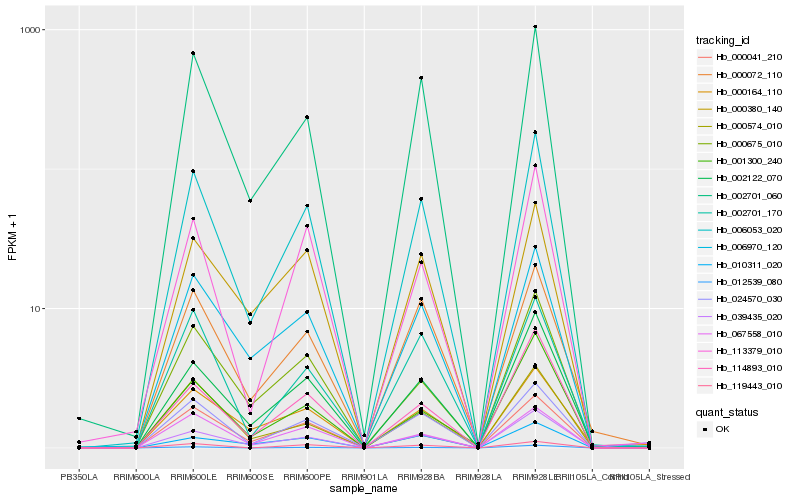
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006053_020 |
0.0 |
- |
- |
CAP [Theobroma cacao] |
| 2 |
Hb_039435_020 |
0.0695339349 |
- |
- |
Lysosomal Pro-X carboxypeptidase, putative [Ricinus communis] |
| 3 |
Hb_002701_060 |
0.0734340107 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002122_070 |
0.0816998478 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_006970_120 |
0.0939641852 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_067558_010 |
0.1009965846 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 7 |
Hb_000072_110 |
0.1083959291 |
- |
- |
PREDICTED: U-box domain-containing protein 35 [Jatropha curcas] |
| 8 |
Hb_114893_010 |
0.1099231337 |
- |
- |
Flavonol 4'-sulfotransferase, putative [Ricinus communis] |
| 9 |
Hb_000675_010 |
0.1100047929 |
- |
- |
PREDICTED: cytokinin dehydrogenase 7 [Jatropha curcas] |
| 10 |
Hb_000380_140 |
0.1114659311 |
- |
- |
PREDICTED: uncharacterized protein LOC105628069 [Jatropha curcas] |
| 11 |
Hb_002701_170 |
0.1136845796 |
- |
- |
PREDICTED: cytochrome P450 85A [Jatropha curcas] |
| 12 |
Hb_113379_010 |
0.1137313034 |
- |
- |
PREDICTED: beta-xylosidase/alpha-L-arabinofuranosidase 1 [Jatropha curcas] |
| 13 |
Hb_000574_010 |
0.1149280484 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_06096 [Jatropha curcas] |
| 14 |
Hb_024570_030 |
0.1152090012 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 15 |
Hb_119443_010 |
0.1188051915 |
- |
- |
hypothetical protein JCGZ_17348 [Jatropha curcas] |
| 16 |
Hb_000164_110 |
0.1219595338 |
transcription factor |
TF Family: C2H2 |
hypothetical protein POPTR_0001s44130g [Populus trichocarpa] |
| 17 |
Hb_001300_240 |
0.1239869815 |
- |
- |
shikimate dehydrogenase, putative [Ricinus communis] |
| 18 |
Hb_012539_080 |
0.1250223878 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 19 |
Hb_000041_210 |
0.1258057226 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 42 [Jatropha curcas] |
| 20 |
Hb_010311_020 |
0.126903617 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |