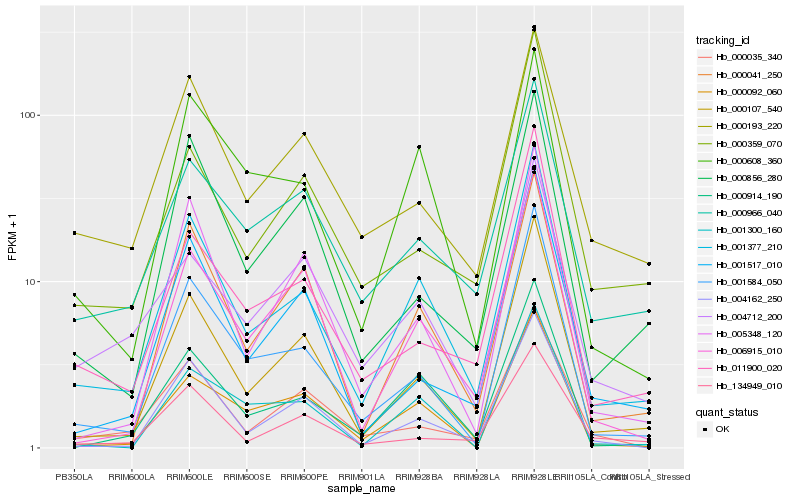
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001377_210 |
0.0 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 2 |
Hb_000914_190 |
0.0980950552 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 3 |
Hb_000107_540 |
0.1016547046 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 4 |
Hb_001584_050 |
0.1102975403 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 5 |
Hb_004162_250 |
0.1279731409 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 6 |
Hb_000608_360 |
0.1329029122 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 7 |
Hb_006915_010 |
0.1351427611 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 8 |
Hb_000092_060 |
0.1373503096 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 9 |
Hb_000041_250 |
0.1375840753 |
- |
- |
PREDICTED: uncharacterized protein LOC105641253 [Jatropha curcas] |
| 10 |
Hb_011900_020 |
0.138000145 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g16390, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001300_160 |
0.1396626041 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 12 |
Hb_000966_040 |
0.139886774 |
- |
- |
hypothetical protein JCGZ_01642 [Jatropha curcas] |
| 13 |
Hb_005348_120 |
0.1412267863 |
- |
- |
PREDICTED: chlorophyll a-b binding protein, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001517_010 |
0.1453223041 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000856_280 |
0.1491714384 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 16 |
Hb_000035_340 |
0.1531362746 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_134949_010 |
0.1540129178 |
- |
- |
PREDICTED: uncharacterized protein LOC105629574 [Jatropha curcas] |
| 18 |
Hb_004712_200 |
0.15502089 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 19 |
Hb_000359_070 |
0.1553325987 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH 2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000193_220 |
0.1553341024 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |