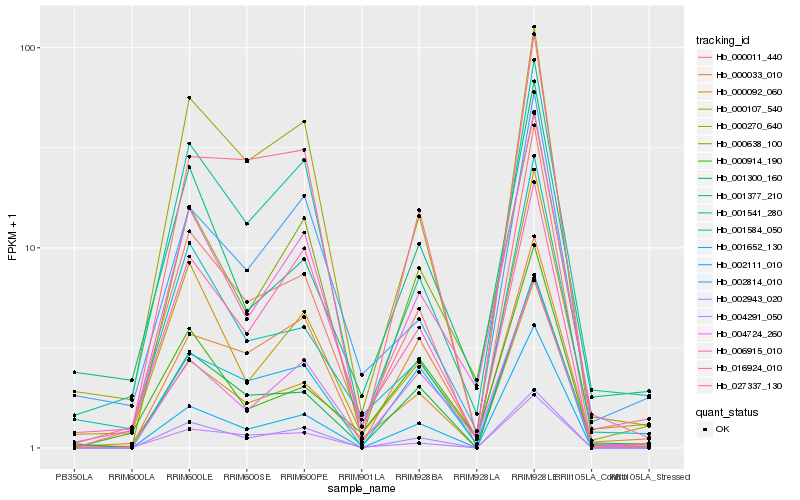
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000092_060 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105646505 [Jatropha curcas] |
| 2 |
Hb_000011_440 |
0.0747692645 |
- |
- |
PREDICTED: GDSL esterase/lipase 1 [Populus euphratica] |
| 3 |
Hb_001300_160 |
0.0864584089 |
- |
- |
PREDICTED: sodium/hydrogen exchanger 2-like [Jatropha curcas] |
| 4 |
Hb_006915_010 |
0.11126423 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_000914_190 |
0.1219009848 |
- |
- |
xanthine dehydrogenase, putative [Ricinus communis] |
| 6 |
Hb_001584_050 |
0.1236224351 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004291_050 |
0.124644609 |
- |
- |
hypothetical protein POPTR_0018s09540g [Populus trichocarpa] |
| 8 |
Hb_002943_020 |
0.1259201975 |
- |
- |
hypothetical protein POPTR_0075s00240g, partial [Populus trichocarpa] |
| 9 |
Hb_000107_540 |
0.1262805697 |
- |
- |
PREDICTED: uncharacterized protein LOC105635335 [Jatropha curcas] |
| 10 |
Hb_004724_260 |
0.127420291 |
- |
- |
PREDICTED: 40S ribosomal protein S14-2-like [Gossypium raimondii] |
| 11 |
Hb_001541_280 |
0.1305423267 |
- |
- |
phosphate transporter [Manihot esculenta] |
| 12 |
Hb_000638_100 |
0.1310061817 |
- |
- |
PREDICTED: histone deacetylase 14 [Jatropha curcas] |
| 13 |
Hb_002814_010 |
0.1323223112 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002111_010 |
0.1323236739 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO2 [Jatropha curcas] |
| 15 |
Hb_000270_640 |
0.1343794442 |
- |
- |
hypothetical protein POPTR_0012s07880g [Populus trichocarpa] |
| 16 |
Hb_001652_130 |
0.1349964996 |
- |
- |
hypothetical protein B456_002G193900 [Gossypium raimondii] |
| 17 |
Hb_027337_130 |
0.1354544849 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 18 |
Hb_001377_210 |
0.1373503096 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 19 |
Hb_000033_010 |
0.1381701397 |
- |
- |
Protein MLO, putative [Ricinus communis] |
| 20 |
Hb_016924_010 |
0.138687759 |
- |
- |
conserved hypothetical protein [Ricinus communis] |