| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
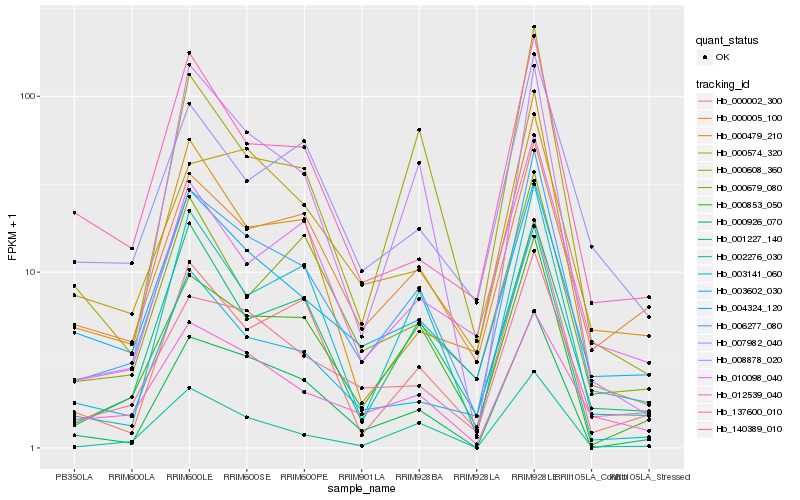
Hb_006277_080 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 2 |
Hb_000853_050 |
0.1040697503 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_140389_010 |
0.1048418494 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 4 |
Hb_000926_070 |
0.1209033631 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002276_030 |
0.1244398774 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 6 |
Hb_003602_030 |
0.1269019876 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 7 |
Hb_003141_060 |
0.1276765426 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 8 |
Hb_001227_140 |
0.1294074944 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 9 |
Hb_000002_300 |
0.1294765444 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 10 |
Hb_000608_360 |
0.1303600944 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 11 |
Hb_004324_120 |
0.1319601518 |
- |
- |
ATP-dependent zinc metalloprotease YME1L1 [Gossypium arboreum] |
| 12 |
Hb_000005_100 |
0.1355574778 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000574_320 |
0.1362649639 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 14 |
Hb_012539_040 |
0.1393529607 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_07765 [Jatropha curcas] |
| 15 |
Hb_010098_040 |
0.1396042314 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 16 |
Hb_007982_040 |
0.1401463016 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 17 |
Hb_137600_010 |
0.1417838595 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 18 |
Hb_000479_210 |
0.1481165043 |
- |
- |
PREDICTED: uncharacterized protein LOC105644126 [Jatropha curcas] |
| 19 |
Hb_008878_020 |
0.1485363811 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 20 |
Hb_000679_080 |
0.1486830783 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |