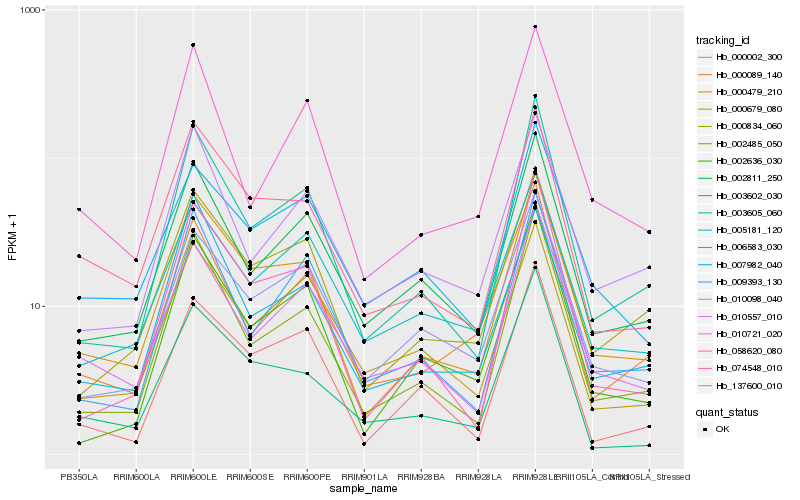
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000479_210 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105644126 [Jatropha curcas] |
| 2 |
Hb_137600_010 |
0.0808762428 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 3 |
Hb_000834_060 |
0.0928376116 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_003605_060 |
0.0944749846 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002485_050 |
0.1043259866 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 6 |
Hb_074548_010 |
0.1092620721 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 7 |
Hb_002811_250 |
0.1165891682 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_010098_040 |
0.117513711 |
- |
- |
PREDICTED: DNA-binding protein SMUBP-2 [Jatropha curcas] |
| 9 |
Hb_002636_030 |
0.121995377 |
- |
- |
hypothetical protein JCGZ_11800 [Jatropha curcas] |
| 10 |
Hb_000002_300 |
0.1225612491 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 11 |
Hb_010721_020 |
0.124051747 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 12 |
Hb_006583_030 |
0.1261251591 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like isoform X4 [Jatropha curcas] |
| 13 |
Hb_003602_030 |
0.1266534993 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 14 |
Hb_007982_040 |
0.1273535632 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 15 |
Hb_010557_010 |
0.131031446 |
- |
- |
PREDICTED: 50S ribosomal protein L3, chloroplastic [Jatropha curcas] |
| 16 |
Hb_005181_120 |
0.1318661757 |
- |
- |
PREDICTED: psbB mRNA maturation factor Mbb1, chloroplastic isoform X1 [Jatropha curcas] |
| 17 |
Hb_009393_130 |
0.1319599357 |
- |
- |
PREDICTED: uncharacterized protein LOC105638053 isoform X4 [Jatropha curcas] |
| 18 |
Hb_058620_080 |
0.1336516966 |
- |
- |
PREDICTED: phosphoglycerate kinase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000089_140 |
0.1352559522 |
- |
- |
PREDICTED: uncharacterized protein LOC105636682 isoform X2 [Jatropha curcas] |
| 20 |
Hb_000679_080 |
0.1369623285 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |