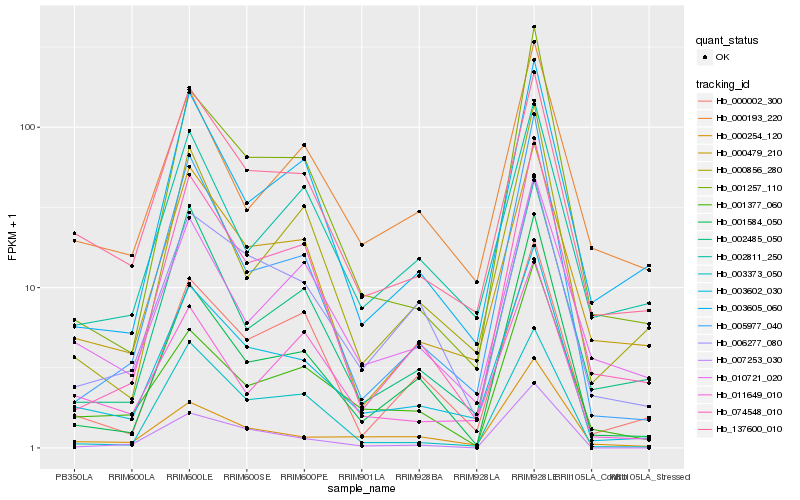
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003602_030 |
0.0 |
- |
- |
PREDICTED: putative pentatricopeptide repeat-containing protein At5g52630 [Jatropha curcas] |
| 2 |
Hb_137600_010 |
0.1046789494 |
- |
- |
50S ribosomal protein L1p, putative [Ricinus communis] |
| 3 |
Hb_000479_210 |
0.1266534993 |
- |
- |
PREDICTED: uncharacterized protein LOC105644126 [Jatropha curcas] |
| 4 |
Hb_006277_080 |
0.1269019876 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 5 |
Hb_001257_110 |
0.1275140296 |
- |
- |
PREDICTED: uncharacterized protein LOC105636396 [Jatropha curcas] |
| 6 |
Hb_000254_120 |
0.1280591269 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_005977_040 |
0.1288323806 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 8 |
Hb_074548_010 |
0.1311218769 |
- |
- |
PREDICTED: 28 kDa ribonucleoprotein, chloroplastic isoform X1 [Jatropha curcas] |
| 9 |
Hb_002485_050 |
0.1365974102 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 12, chloroplastic [Jatropha curcas] |
| 10 |
Hb_003605_060 |
0.1393220491 |
- |
- |
PREDICTED: ATP synthase subunit delta, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001377_060 |
0.1409425663 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At4g01030, mitochondrial [Jatropha curcas] |
| 12 |
Hb_000002_300 |
0.1437074713 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 13 |
Hb_000856_280 |
0.1440602997 |
- |
- |
PREDICTED: uncharacterized protein LOC105640491 [Jatropha curcas] |
| 14 |
Hb_011649_010 |
0.1443727254 |
transcription factor |
TF Family: Pseudo ARR-B |
sensory transduction histidine kinase, putative [Ricinus communis] |
| 15 |
Hb_007253_030 |
0.1449155335 |
- |
- |
PREDICTED: 33 kDa ribonucleoprotein, chloroplastic-like [Populus euphratica] |
| 16 |
Hb_000193_220 |
0.1466677409 |
- |
- |
PREDICTED: ribulose-phosphate 3-epimerase, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002811_250 |
0.1484016933 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 3, chloroplastic [Jatropha curcas] |
| 18 |
Hb_010721_020 |
0.1485987987 |
- |
- |
PREDICTED: photosynthetic NDH subunit of lumenal location 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_001584_050 |
0.1490490096 |
- |
- |
PREDICTED: NHL repeat-containing protein 2 isoform X1 [Jatropha curcas] |
| 20 |
Hb_003373_050 |
0.14959556 |
- |
- |
receptor protein kinase zmpk1, putative [Ricinus communis] |