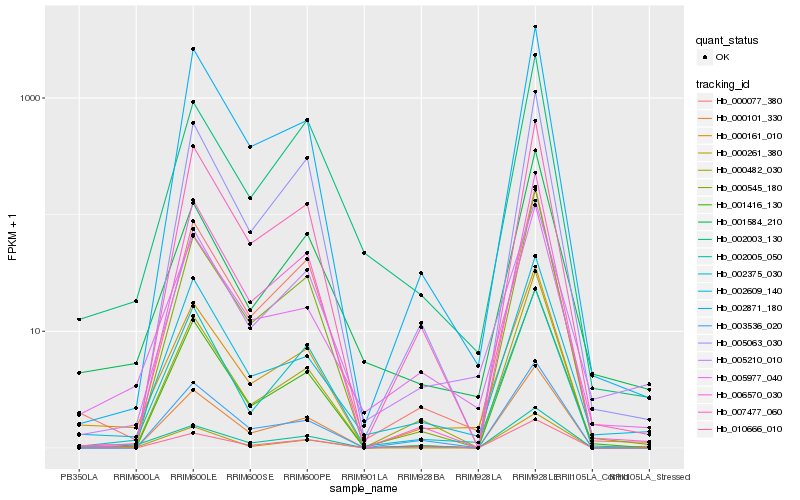
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000161_010 |
0.0 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000077_380 |
0.0927972502 |
- |
- |
PREDICTED: protochlorophyllide reductase [Jatropha curcas] |
| 3 |
Hb_005977_040 |
0.1008242963 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 4 |
Hb_000101_330 |
0.1155424537 |
- |
- |
- |
| 5 |
Hb_002609_140 |
0.1209107133 |
- |
- |
PREDICTED: sodium-dependent phosphate transport protein 1, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_001584_210 |
0.1210822618 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa b, chloroplastic [Jatropha curcas] |
| 7 |
Hb_007477_060 |
0.1216496365 |
- |
- |
unknown [Medicago truncatula] |
| 8 |
Hb_000545_180 |
0.1237429454 |
- |
- |
Photosystem II stability/assembly factor HCF136, chloroplast precursor, putative [Ricinus communis] |
| 9 |
Hb_000482_030 |
0.1242822335 |
transcription factor |
TF Family: C2H2 |
PREDICTED: zinc finger protein NUTCRACKER-like isoform X1 [Populus euphratica] |
| 10 |
Hb_006570_030 |
0.1249153866 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002871_180 |
0.1256622648 |
- |
- |
photosystem II 10 kDa polypeptide, chloroplastic [Jatropha curcas] |
| 12 |
Hb_002005_050 |
0.125753112 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 13 |
Hb_005210_010 |
0.1267212026 |
- |
- |
PREDICTED: uncharacterized protein LOC105638163 [Jatropha curcas] |
| 14 |
Hb_010666_010 |
0.127917854 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_002375_030 |
0.1280495907 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 16 |
Hb_002003_130 |
0.1281719159 |
- |
- |
hypothetical protein CISIN_1g027843mg [Citrus sinensis] |
| 17 |
Hb_003536_020 |
0.1296629822 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 18 |
Hb_001416_130 |
0.1299089274 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000261_380 |
0.1307228884 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_1019500 [Ricinus communis] |
| 20 |
Hb_005063_030 |
0.1311168983 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |