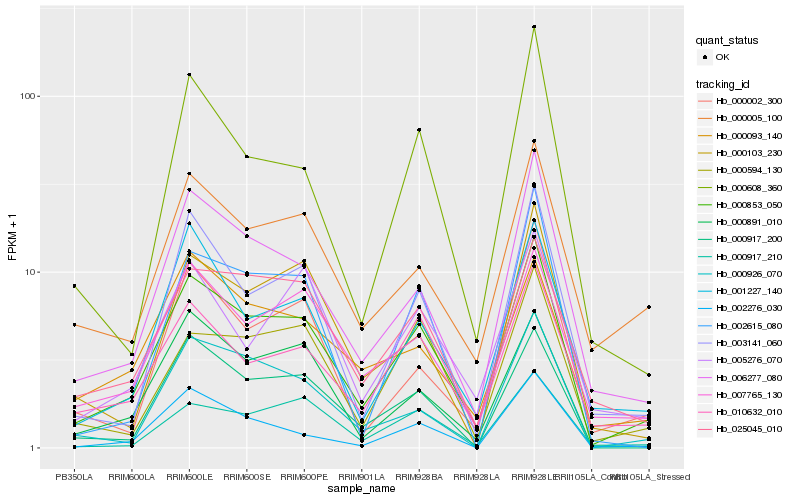
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000853_050 |
0.0 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_006277_080 |
0.1040697503 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 3 |
Hb_010632_010 |
0.125527147 |
- |
- |
PREDICTED: bifunctional dethiobiotin synthetase/7,8-diamino-pelargonic acid aminotransferase, mitochondrial [Jatropha curcas] |
| 4 |
Hb_007765_130 |
0.1273306212 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 5 |
Hb_003141_060 |
0.1293991129 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 6 |
Hb_001227_140 |
0.1359687597 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 7 |
Hb_025045_010 |
0.1362864321 |
- |
- |
catalytic, putative [Ricinus communis] |
| 8 |
Hb_000005_100 |
0.1372535358 |
- |
- |
PREDICTED: pheophorbide a oxygenase, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000917_200 |
0.1377362914 |
- |
- |
hypothetical protein PRUPE_ppa010555mg [Prunus persica] |
| 10 |
Hb_000917_210 |
0.1402790834 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000608_360 |
0.1412951321 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 12 |
Hb_000891_010 |
0.1428676185 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000594_130 |
0.1456959905 |
- |
- |
serine-threonine protein kinase, plant-type, putative [Ricinus communis] |
| 14 |
Hb_000926_070 |
0.1467354489 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000002_300 |
0.1507573355 |
- |
- |
Desacetoxyvindoline 4-hydroxylase, putative [Ricinus communis] |
| 16 |
Hb_000093_140 |
0.1538576741 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_002276_030 |
0.1541575905 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 18 |
Hb_002615_080 |
0.1543958612 |
- |
- |
PREDICTED: non-specific phospholipase C2 [Jatropha curcas] |
| 19 |
Hb_000103_230 |
0.154505513 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |
| 20 |
Hb_005276_070 |
0.1555545899 |
- |
- |
PREDICTED: RAN GTPase-activating protein 1 [Jatropha curcas] |