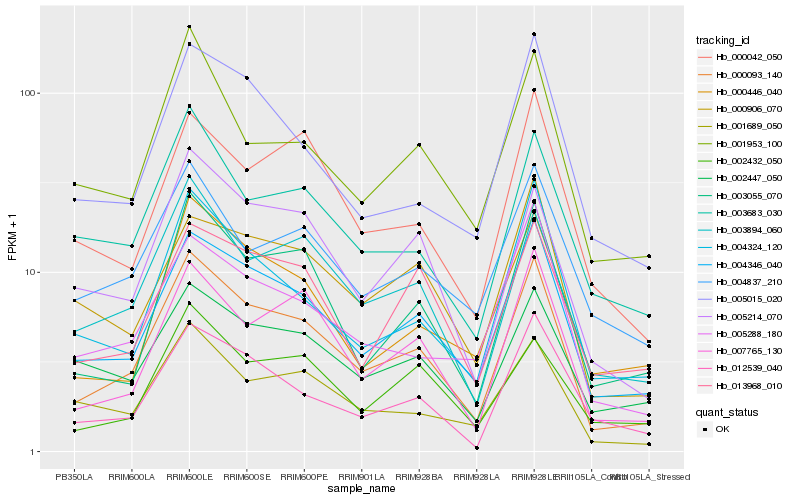
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000093_140 |
0.0 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_004346_040 |
0.0802061267 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 3 |
Hb_003894_060 |
0.1057667419 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_003055_070 |
0.1069545927 |
- |
- |
PREDICTED: katanin p80 WD40 repeat-containing subunit B1 homolog isoform X1 [Jatropha curcas] |
| 5 |
Hb_007765_130 |
0.11006291 |
- |
- |
PREDICTED: UPF0503 protein At3g09070, chloroplastic [Jatropha curcas] |
| 6 |
Hb_005214_070 |
0.1172281677 |
- |
- |
PREDICTED: fatty acid amide hydrolase [Jatropha curcas] |
| 7 |
Hb_004324_120 |
0.1176300487 |
- |
- |
ATP-dependent zinc metalloprotease YME1L1 [Gossypium arboreum] |
| 8 |
Hb_012539_040 |
0.1198324709 |
transcription factor |
TF Family: CPP |
hypothetical protein JCGZ_07765 [Jatropha curcas] |
| 9 |
Hb_000446_040 |
0.1225154756 |
- |
- |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 10 |
Hb_001689_050 |
0.1257116219 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 11 |
Hb_005015_020 |
0.1261978648 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 12 |
Hb_003683_030 |
0.1285939465 |
- |
- |
PREDICTED: uridine-cytidine kinase C isoform X1 [Jatropha curcas] |
| 13 |
Hb_001953_100 |
0.128681253 |
- |
- |
PREDICTED: glutamate-1-semialdehyde 2,1-aminomutase 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_005288_180 |
0.1348804165 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_000042_050 |
0.135346157 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 16 |
Hb_013968_010 |
0.1353675233 |
- |
- |
PREDICTED: U4/U6 small nuclear ribonucleoprotein Prp3 [Jatropha curcas] |
| 17 |
Hb_002447_050 |
0.136648111 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 18 |
Hb_004837_210 |
0.1366498393 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 19 |
Hb_002432_050 |
0.1399065059 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 20 |
Hb_000906_070 |
0.1400706116 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |