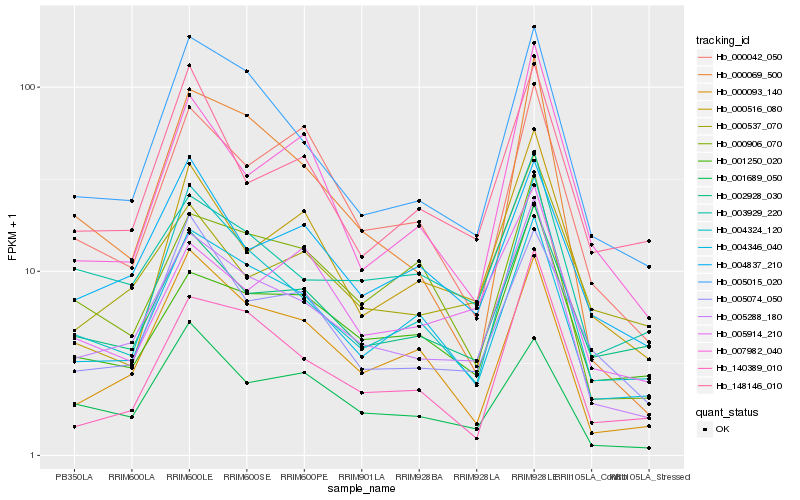
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005288_180 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |
| 2 |
Hb_005015_020 |
0.1022091269 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_004346_040 |
0.1093486954 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 4 |
Hb_004324_120 |
0.1109176461 |
- |
- |
ATP-dependent zinc metalloprotease YME1L1 [Gossypium arboreum] |
| 5 |
Hb_001250_020 |
0.1259571917 |
- |
- |
PREDICTED: CRS2-associated factor 2, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003929_220 |
0.1279763483 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_002928_030 |
0.1288945983 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 8 |
Hb_140389_010 |
0.1303121951 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 9 |
Hb_000093_140 |
0.1348804165 |
- |
- |
PREDICTED: fructokinase-like 2, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000069_500 |
0.1370507191 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_000906_070 |
0.1395798929 |
- |
- |
PREDICTED: protein SCAR2-like isoform X1 [Jatropha curcas] |
| 12 |
Hb_004837_210 |
0.1412406238 |
- |
- |
PREDICTED: calcium-dependent protein kinase 8-like [Jatropha curcas] |
| 13 |
Hb_000042_050 |
0.1426648163 |
- |
- |
PREDICTED: uncharacterized protein LOC105632791 isoform X1 [Jatropha curcas] |
| 14 |
Hb_007982_040 |
0.1441972192 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase At1g79600, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000537_070 |
0.1444218787 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 16 |
Hb_001689_050 |
0.1454190999 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Prunus mume] |
| 17 |
Hb_148146_010 |
0.1463635218 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 18 |
Hb_000516_080 |
0.1466097895 |
- |
- |
hypothetical protein POPTR_0010s24810g [Populus trichocarpa] |
| 19 |
Hb_005074_050 |
0.1482085881 |
- |
- |
PREDICTED: myosin-6-like [Jatropha curcas] |
| 20 |
Hb_005914_210 |
0.1493180024 |
- |
- |
PREDICTED: uncharacterized protein LOC105638411 [Jatropha curcas] |