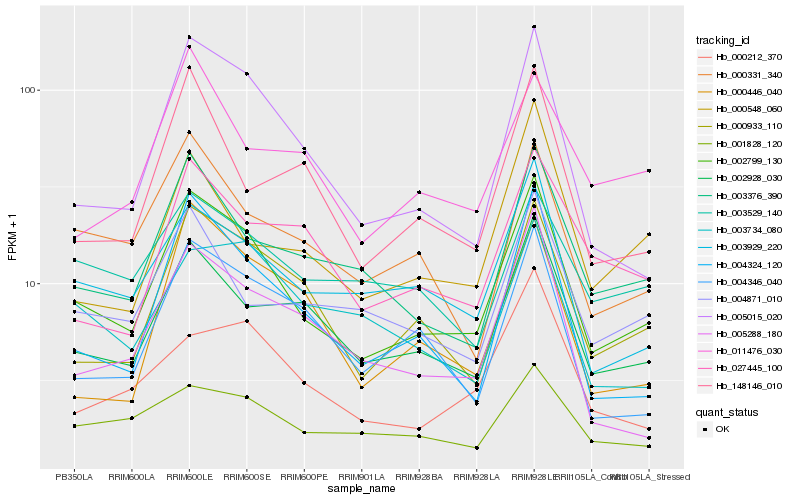
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002799_130 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105638514 [Jatropha curcas] |
| 2 |
Hb_005015_020 |
0.1168821183 |
desease resistance |
Gene Name: ABC_tran |
ABC transporter family protein [Hevea brasiliensis] |
| 3 |
Hb_000933_110 |
0.118699657 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 3 [Jatropha curcas] |
| 4 |
Hb_002928_030 |
0.1218787049 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 5 |
Hb_003376_390 |
0.128534873 |
- |
- |
PREDICTED: 29 kDa ribonucleoprotein A, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003929_220 |
0.1288998886 |
- |
- |
pentatricopeptide repeat-containing protein, putative [Ricinus communis] |
| 7 |
Hb_004324_120 |
0.1289233075 |
- |
- |
ATP-dependent zinc metalloprotease YME1L1 [Gossypium arboreum] |
| 8 |
Hb_000331_340 |
0.1378161219 |
- |
- |
PREDICTED: uncharacterized protein LOC105640234 [Jatropha curcas] |
| 9 |
Hb_004871_010 |
0.1459600051 |
- |
- |
RNA polymerase sigma factor rpoD1, putative [Ricinus communis] |
| 10 |
Hb_004346_040 |
0.1492155921 |
- |
- |
hypothetical protein POPTR_0016s06630g [Populus trichocarpa] |
| 11 |
Hb_001828_120 |
0.1498812893 |
- |
- |
PREDICTED: uncharacterized protein LOC105140167 [Populus euphratica] |
| 12 |
Hb_000212_370 |
0.1516392207 |
- |
- |
thioredoxin f-type, putative [Ricinus communis] |
| 13 |
Hb_005288_180 |
0.1517755293 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g04760, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_148146_010 |
0.1517998363 |
- |
- |
coproporphyrinogen III oxidase [Ziziphus jujuba] |
| 15 |
Hb_027445_100 |
0.1525318077 |
- |
- |
PREDICTED: probable monodehydroascorbate reductase, cytoplasmic isoform 2 [Jatropha curcas] |
| 16 |
Hb_003529_140 |
0.1555086589 |
- |
- |
PREDICTED: nifU-like protein 2, chloroplastic [Jatropha curcas] |
| 17 |
Hb_003734_080 |
0.1579806545 |
- |
- |
myosin vIII, putative [Ricinus communis] |
| 18 |
Hb_000446_040 |
0.1580765788 |
- |
- |
PREDICTED: uncharacterized protein ycf45 [Jatropha curcas] |
| 19 |
Hb_000548_060 |
0.1593035571 |
- |
- |
PREDICTED: fruit protein pKIWI502 [Jatropha curcas] |
| 20 |
Hb_011476_030 |
0.159523601 |
- |
- |
PREDICTED: uncharacterized protein LOC105632352 [Jatropha curcas] |