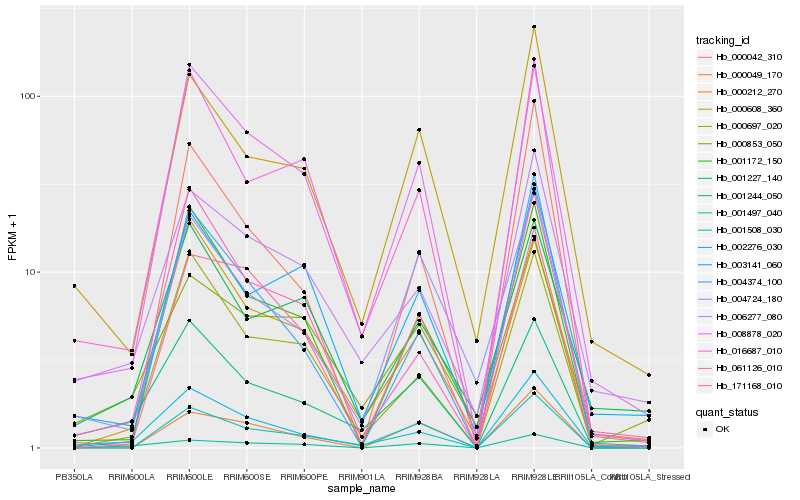
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002276_030 |
0.0 |
- |
- |
PREDICTED: putative U-box domain-containing protein 42 [Jatropha curcas] |
| 2 |
Hb_008878_020 |
0.1192029393 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 3 |
Hb_001244_050 |
0.1208185579 |
- |
- |
PREDICTED: uncharacterized protein LOC105643334 [Jatropha curcas] |
| 4 |
Hb_006277_080 |
0.1244398774 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 5 |
Hb_000608_360 |
0.1325146592 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 6 |
Hb_001508_030 |
0.1398638382 |
- |
- |
hypothetical protein VITISV_029808 [Vitis vinifera] |
| 7 |
Hb_001172_150 |
0.142255587 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 8 |
Hb_004374_100 |
0.1458645188 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |
| 9 |
Hb_016687_010 |
0.1475495682 |
- |
- |
PREDICTED: uncharacterized protein LOC105637401 [Jatropha curcas] |
| 10 |
Hb_000697_020 |
0.1484213427 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_000042_310 |
0.1512254992 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 12 |
Hb_000212_270 |
0.1522075424 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 13 |
Hb_000853_050 |
0.1541575905 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_003141_060 |
0.1562385365 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 15 |
Hb_061126_010 |
0.1566974397 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 16 |
Hb_004724_180 |
0.1585249721 |
- |
- |
PREDICTED: protein FANTASTIC FOUR 1 [Jatropha curcas] |
| 17 |
Hb_001227_140 |
0.1590308475 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 18 |
Hb_001497_040 |
0.1622281806 |
- |
- |
hypothetical protein POPTR_0008s219701g, partial [Populus trichocarpa] |
| 19 |
Hb_171168_010 |
0.1652276118 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 20 |
Hb_000049_170 |
0.1688769151 |
- |
- |
conserved hypothetical protein [Ricinus communis] |