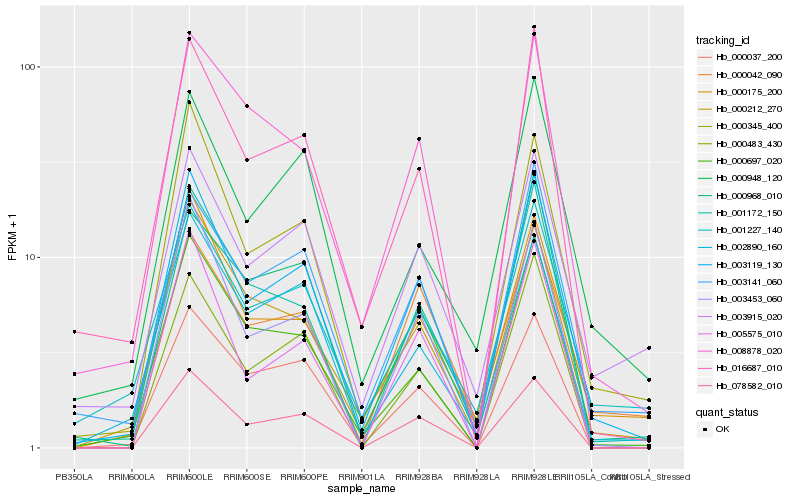
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003119_130 |
0.0 |
- |
- |
hypothetical protein POPTR_0017s07770g [Populus trichocarpa] |
| 2 |
Hb_078582_010 |
0.097242901 |
- |
- |
hypothetical protein POPTR_0005s01620g [Populus trichocarpa] |
| 3 |
Hb_016687_010 |
0.0982920385 |
- |
- |
PREDICTED: uncharacterized protein LOC105637401 [Jatropha curcas] |
| 4 |
Hb_003453_060 |
0.0991090404 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 5 |
Hb_000175_200 |
0.0991406492 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001172_150 |
0.0992631085 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 7 |
Hb_005575_010 |
0.1041286717 |
- |
- |
hypothetical protein POPTR_0004s02560g [Populus trichocarpa] |
| 8 |
Hb_000212_270 |
0.1050499474 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 9 |
Hb_000697_020 |
0.1058593866 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 10 |
Hb_003141_060 |
0.106288939 |
transcription factor |
TF Family: TRAF |
PREDICTED: ETO1-like protein 1 [Jatropha curcas] |
| 11 |
Hb_000037_200 |
0.1117920213 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 12 |
Hb_002890_160 |
0.1120109834 |
- |
- |
PREDICTED: protein root UVB sensitive 6 isoform X1 [Jatropha curcas] |
| 13 |
Hb_000483_430 |
0.1146912275 |
- |
- |
- |
| 14 |
Hb_008878_020 |
0.1203536653 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 15 |
Hb_003915_020 |
0.1204604438 |
- |
- |
PREDICTED: outer envelope pore protein 37, chloroplastic [Jatropha curcas] |
| 16 |
Hb_001227_140 |
0.1226119994 |
- |
- |
PREDICTED: protein ROS1 [Jatropha curcas] |
| 17 |
Hb_000948_120 |
0.1257452571 |
- |
- |
PREDICTED: uncharacterized protein LOC105634688 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000042_090 |
0.1285715131 |
desease resistance |
Gene Name: NB-ARC |
Disease resistance protein RPS2, putative [Ricinus communis] |
| 19 |
Hb_000968_010 |
0.1289030906 |
- |
- |
PREDICTED: glutamate receptor 3.7 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000345_400 |
0.1296659509 |
- |
- |
conserved hypothetical protein [Ricinus communis] |