| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
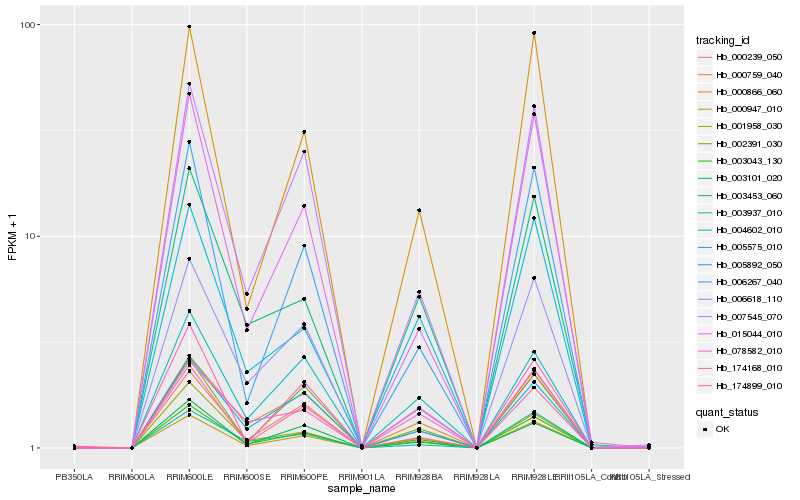
Hb_003043_130 |
0.0 |
transcription factor |
TF Family: ERF |
PREDICTED: ethylene-responsive transcription factor CRF2-like [Jatropha curcas] |
| 2 |
Hb_000947_010 |
0.0616145762 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: putative disease resistance protein RGA4 [Jatropha curcas] |
| 3 |
Hb_000866_060 |
0.088244129 |
- |
- |
putative glycerol-3-phosphate acyltransferase [Vernicia fordii] |
| 4 |
Hb_003453_060 |
0.0895818273 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 5 |
Hb_005575_010 |
0.0922154328 |
- |
- |
hypothetical protein POPTR_0004s02560g [Populus trichocarpa] |
| 6 |
Hb_000759_040 |
0.0941480345 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At1g34300 [Jatropha curcas] |
| 7 |
Hb_000239_050 |
0.094414871 |
- |
- |
PREDICTED: cyclin-D5-1-like [Jatropha curcas] |
| 8 |
Hb_174168_010 |
0.0944880365 |
- |
- |
hypothetical protein JCGZ_06105 [Jatropha curcas] |
| 9 |
Hb_007545_070 |
0.0958395001 |
transcription factor |
TF Family: MYB |
PREDICTED: myb-related protein 306 [Populus euphratica] |
| 10 |
Hb_006267_040 |
0.1025645968 |
- |
- |
PREDICTED: ABC transporter G family member 32 isoform X1 [Jatropha curcas] |
| 11 |
Hb_015044_010 |
0.1027562544 |
- |
- |
hypothetical protein JCGZ_25130 [Jatropha curcas] |
| 12 |
Hb_005892_050 |
0.1037186158 |
- |
- |
PREDICTED: uncharacterized protein LOC105632175 [Jatropha curcas] |
| 13 |
Hb_006618_110 |
0.1055622777 |
- |
- |
- |
| 14 |
Hb_174899_010 |
0.1060063993 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180 [Jatropha curcas] |
| 15 |
Hb_004602_010 |
0.1065394702 |
- |
- |
PREDICTED: uncharacterized protein LOC105042236 isoform X2 [Elaeis guineensis] |
| 16 |
Hb_003101_020 |
0.1082215481 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC transcription factor NAM-B2 [Jatropha curcas] |
| 17 |
Hb_078582_010 |
0.1108352346 |
- |
- |
hypothetical protein POPTR_0005s01620g [Populus trichocarpa] |
| 18 |
Hb_001958_030 |
0.1130821913 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 19 |
Hb_003937_010 |
0.1168819104 |
- |
- |
hypothetical protein JCGZ_20930 [Jatropha curcas] |
| 20 |
Hb_002391_030 |
0.1169805333 |
- |
- |
hypothetical protein CISIN_1g020187mg [Citrus sinensis] |