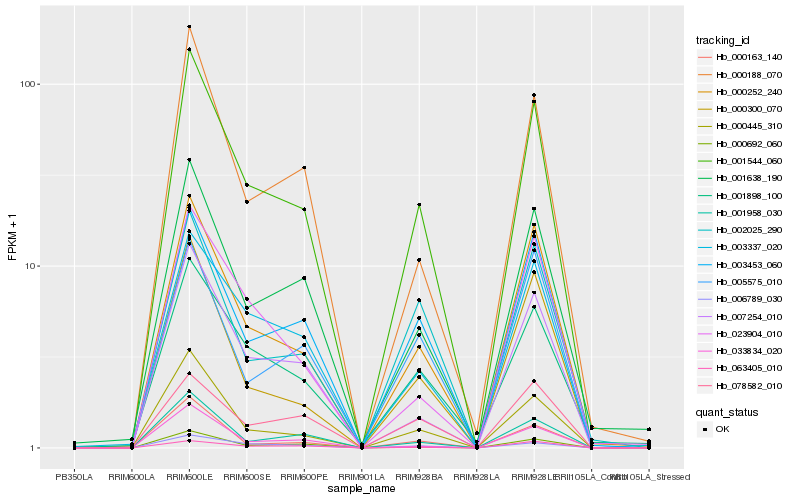
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001544_060 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000252_240 |
0.0767245409 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X2 [Gossypium raimondii] |
| 3 |
Hb_000445_310 |
0.0837306777 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 4 |
Hb_003453_060 |
0.0931378148 |
- |
- |
PREDICTED: nudix hydrolase 13, mitochondrial [Jatropha curcas] |
| 5 |
Hb_001898_100 |
0.1005763045 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 6 |
Hb_003337_020 |
0.1036066222 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 7 |
Hb_001638_190 |
0.105842913 |
- |
- |
PREDICTED: protein IQ-DOMAIN 1-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_007254_010 |
0.1064792489 |
- |
- |
hypothetical protein JCGZ_25214 [Jatropha curcas] |
| 9 |
Hb_006789_030 |
0.1086280559 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 10 |
Hb_000692_060 |
0.1120892308 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 11 |
Hb_001958_030 |
0.1121462098 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4-like [Jatropha curcas] |
| 12 |
Hb_000188_070 |
0.1156513132 |
- |
- |
calcineurin-like phosphoesterase [Manihot esculenta] |
| 13 |
Hb_000300_070 |
0.1182104795 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 14 |
Hb_002025_290 |
0.1225306125 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000163_140 |
0.1235081218 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 16 |
Hb_033834_020 |
0.128734816 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_023904_010 |
0.131957611 |
- |
- |
- |
| 18 |
Hb_063405_010 |
0.1380516463 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 19 |
Hb_005575_010 |
0.1382338492 |
- |
- |
hypothetical protein POPTR_0004s02560g [Populus trichocarpa] |
| 20 |
Hb_078582_010 |
0.1389799104 |
- |
- |
hypothetical protein POPTR_0005s01620g [Populus trichocarpa] |