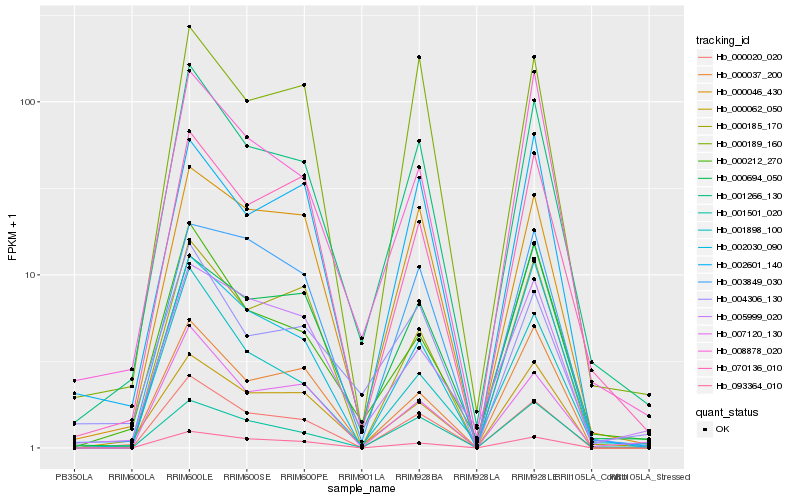
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001266_130 |
0.0 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 2 |
Hb_000020_020 |
0.1025182949 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 3 |
Hb_008878_020 |
0.1105956265 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 4 |
Hb_000212_270 |
0.1124167268 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 5 |
Hb_000062_050 |
0.1141414442 |
- |
- |
protein binding protein, putative [Ricinus communis] |
| 6 |
Hb_001501_020 |
0.1144314398 |
- |
- |
PREDICTED: receptor-like protein kinase [Jatropha curcas] |
| 7 |
Hb_000046_430 |
0.1158953332 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 8 |
Hb_070136_010 |
0.1209824453 |
- |
- |
TRANSPARENT TESTA 12 protein, putative [Ricinus communis] |
| 9 |
Hb_000189_160 |
0.1215147191 |
- |
- |
PREDICTED: GDP-mannose 3,5-epimerase 2 [Eucalyptus grandis] |
| 10 |
Hb_004306_130 |
0.1226965714 |
- |
- |
PREDICTED: uncharacterized protein LOC105644385 isoform X1 [Jatropha curcas] |
| 11 |
Hb_001898_100 |
0.1228413724 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 12 |
Hb_002030_090 |
0.1240700413 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 13 |
Hb_007120_130 |
0.1256450795 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 14 |
Hb_000694_050 |
0.1289338143 |
- |
- |
Nuclease PA3, putative [Ricinus communis] |
| 15 |
Hb_000037_200 |
0.1311783885 |
- |
- |
RNA binding protein, putative [Ricinus communis] |
| 16 |
Hb_000185_170 |
0.1315535036 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 17 |
Hb_093364_010 |
0.1320054379 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 18 |
Hb_005999_020 |
0.1370060048 |
transcription factor |
TF Family: C2H2 |
PREDICTED: protein SENSITIVE TO PROTON RHIZOTOXICITY 1-like [Jatropha curcas] |
| 19 |
Hb_003849_030 |
0.1410866641 |
- |
- |
hypothetical protein AALP_AA5G029900 [Arabis alpina] |
| 20 |
Hb_002601_140 |
0.1411288043 |
- |
- |
signal transducer, putative [Ricinus communis] |