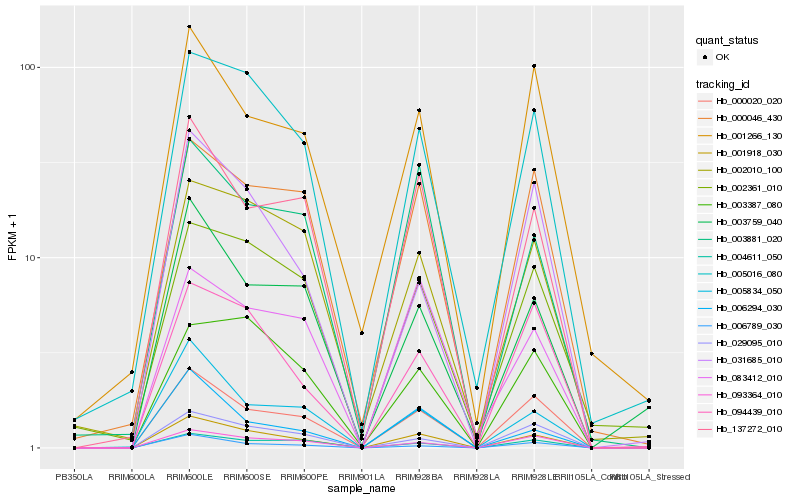
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001918_030 |
0.0 |
- |
- |
- |
| 2 |
Hb_000020_020 |
0.0957351194 |
- |
- |
hypothetical protein JCGZ_06491 [Jatropha curcas] |
| 3 |
Hb_005016_080 |
0.1115256719 |
- |
- |
PREDICTED: neurofilament heavy polypeptide-like [Jatropha curcas] |
| 4 |
Hb_137272_010 |
0.1209350608 |
- |
- |
PREDICTED: cysteine-rich receptor-like protein kinase 10 [Jatropha curcas] |
| 5 |
Hb_093364_010 |
0.1255279236 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 6 |
Hb_029095_010 |
0.1340566764 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 7 |
Hb_094439_010 |
0.1342905176 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 8 |
Hb_004611_050 |
0.1359162647 |
- |
- |
PREDICTED: uncharacterized protein LOC105628744 [Jatropha curcas] |
| 9 |
Hb_005834_050 |
0.1369003607 |
- |
- |
PREDICTED: cationic amino acid transporter 1-like [Jatropha curcas] |
| 10 |
Hb_003881_020 |
0.1432137983 |
- |
- |
PREDICTED: stellacyanin-like [Jatropha curcas] |
| 11 |
Hb_006789_030 |
0.14413045 |
- |
- |
PREDICTED: wall-associated receptor kinase-like 22 [Prunus mume] |
| 12 |
Hb_000046_430 |
0.1446957229 |
- |
- |
hypothetical protein JCGZ_02013 [Jatropha curcas] |
| 13 |
Hb_002010_100 |
0.1492615238 |
- |
- |
PREDICTED: uncharacterized protein LOC105633355 [Jatropha curcas] |
| 14 |
Hb_003387_080 |
0.1493334769 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_006294_030 |
0.1503747351 |
- |
- |
alcohol dehydrogenase, putative [Ricinus communis] |
| 16 |
Hb_003759_040 |
0.1510958397 |
- |
- |
PREDICTED: aldo-keto reductase family 4 member C9-like [Jatropha curcas] |
| 17 |
Hb_002361_010 |
0.1530166922 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 18 |
Hb_001266_130 |
0.1562377586 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase-like [Jatropha curcas] |
| 19 |
Hb_083412_010 |
0.1565272832 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 20 |
Hb_031685_010 |
0.15663712 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |