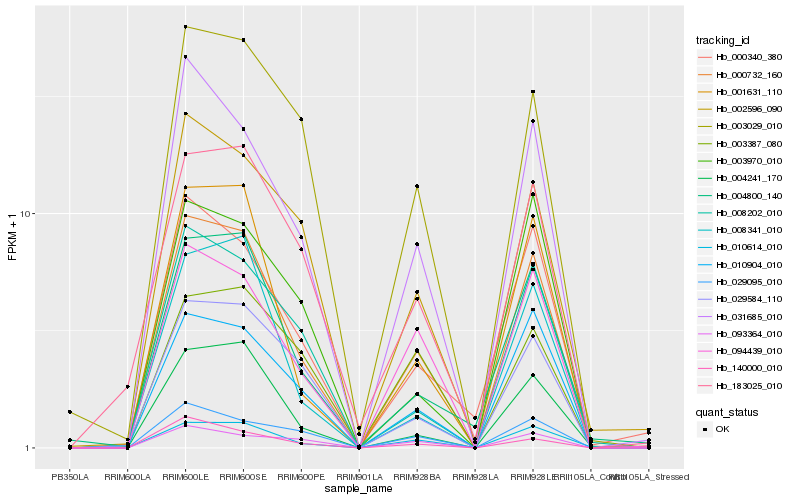
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000732_160 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 2 |
Hb_010614_010 |
0.0931118822 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 3 |
Hb_031685_010 |
0.0970292687 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 4 |
Hb_094439_010 |
0.097704039 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 5 |
Hb_010904_010 |
0.1026805473 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 6 |
Hb_001631_110 |
0.1030086872 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: gamma-glutamyltranspeptidase 1-like [Populus euphratica] |
| 7 |
Hb_004241_170 |
0.1064835243 |
- |
- |
PREDICTED: uncharacterized protein LOC105633220 [Jatropha curcas] |
| 8 |
Hb_029584_110 |
0.1252864868 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 9 |
Hb_029095_010 |
0.1266957422 |
desease resistance |
Gene Name: NB-ARC |
hypothetical protein JCGZ_21060 [Jatropha curcas] |
| 10 |
Hb_003029_010 |
0.1284417031 |
transcription factor |
TF Family: WRKY |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004800_140 |
0.1287413691 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 12 |
Hb_002596_090 |
0.1295877068 |
- |
- |
PREDICTED: calcium-transporting ATPase 4, plasma membrane-type-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000340_380 |
0.1296934776 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 14 |
Hb_008202_010 |
0.1297640356 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_008341_010 |
0.1306147493 |
transcription factor |
TF Family: Orphans |
PREDICTED: zinc finger protein HD1-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_183025_010 |
0.1346185488 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 17 |
Hb_003970_010 |
0.1462295968 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_140000_010 |
0.147476033 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_093364_010 |
0.1489281491 |
- |
- |
PREDICTED: uncharacterized protein LOC105636199 [Jatropha curcas] |
| 20 |
Hb_003387_080 |
0.1557490166 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |