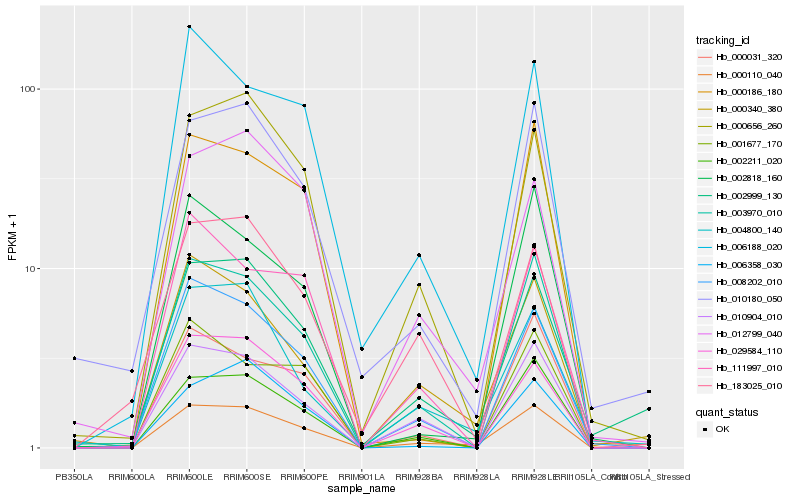
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000110_040 |
0.0 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase CVP2 [Jatropha curcas] |
| 2 |
Hb_003970_010 |
0.103876138 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 3 |
Hb_002211_020 |
0.1137776476 |
transcription factor |
TF Family: C2C2-GATA |
PREDICTED: GATA transcription factor 5-like [Jatropha curcas] |
| 4 |
Hb_010904_010 |
0.1189601124 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 5 |
Hb_008202_010 |
0.1269528272 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000656_260 |
0.1283786634 |
- |
- |
PREDICTED: uncharacterized protein LOC105631376 [Jatropha curcas] |
| 7 |
Hb_000340_380 |
0.1314346063 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 8 |
Hb_029584_110 |
0.1320641318 |
transcription factor |
TF Family: MYB-related |
PREDICTED: myb-like protein J [Jatropha curcas] |
| 9 |
Hb_002999_130 |
0.13729081 |
transcription factor |
TF Family: C2C2-Dof |
hypothetical protein POPTR_0008s08740g [Populus trichocarpa] |
| 10 |
Hb_004800_140 |
0.140631961 |
- |
- |
PREDICTED: uncharacterized protein LOC105633940 isoform X1 [Jatropha curcas] |
| 11 |
Hb_012799_040 |
0.1413824678 |
- |
- |
PREDICTED: transmembrane ascorbate ferrireductase 1 [Jatropha curcas] |
| 12 |
Hb_010180_050 |
0.1453918439 |
- |
- |
PREDICTED: CBL-interacting protein kinase 2 [Jatropha curcas] |
| 13 |
Hb_006188_020 |
0.1465084635 |
- |
- |
PREDICTED: uncharacterized protein LOC105646403 [Jatropha curcas] |
| 14 |
Hb_000031_320 |
0.1474837847 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 15 |
Hb_000186_180 |
0.1523167032 |
- |
- |
PREDICTED: uncharacterized protein LOC105649287 [Jatropha curcas] |
| 16 |
Hb_183025_010 |
0.1564817994 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RMA1H1 [Jatropha curcas] |
| 17 |
Hb_006358_030 |
0.1598849438 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 18 |
Hb_002818_160 |
0.1602245521 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 19 |
Hb_001677_170 |
0.1610067926 |
- |
- |
hypothetical protein M569_03661, partial [Genlisea aurea] |
| 20 |
Hb_111997_010 |
0.1617492355 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase RLK1 [Vitis vinifera] |