| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
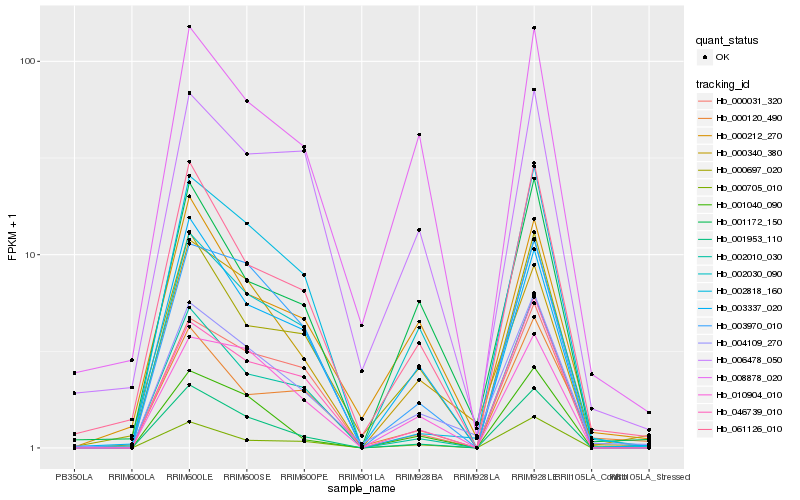
Hb_004109_270 |
0.0 |
transcription factor |
TF Family: NAC |
transcription activator NAC1 family protein [Populus trichocarpa] |
| 2 |
Hb_000340_380 |
0.1221433988 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 3 |
Hb_001172_150 |
0.1233818735 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 4 |
Hb_061126_010 |
0.124466324 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_000697_020 |
0.1325917269 |
- |
- |
Zinc-binding dehydrogenase family protein isoform 1 [Theobroma cacao] |
| 6 |
Hb_001953_110 |
0.1334415765 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 7 |
Hb_008878_020 |
0.137930017 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 8 |
Hb_046739_010 |
0.1389080411 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 9 |
Hb_002010_030 |
0.141391002 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 10 |
Hb_000120_490 |
0.1428195617 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like [Jatropha curcas] |
| 11 |
Hb_000212_270 |
0.1435313671 |
- |
- |
calreticulin, putative [Ricinus communis] |
| 12 |
Hb_003970_010 |
0.1458228723 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 13 |
Hb_001040_090 |
0.1468768192 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |
| 14 |
Hb_010904_010 |
0.1468985064 |
- |
- |
PREDICTED: probable receptor-like protein kinase At5g39030 isoform X2 [Jatropha curcas] |
| 15 |
Hb_002818_160 |
0.1472023264 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 16 |
Hb_000705_010 |
0.1475351556 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 17 |
Hb_000031_320 |
0.1477818367 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 18 |
Hb_002030_090 |
0.1478751588 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 19 |
Hb_003337_020 |
0.1495053901 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |
| 20 |
Hb_006478_050 |
0.1509946768 |
- |
- |
PREDICTED: serine/threonine-protein kinase HT1 isoform X1 [Jatropha curcas] |