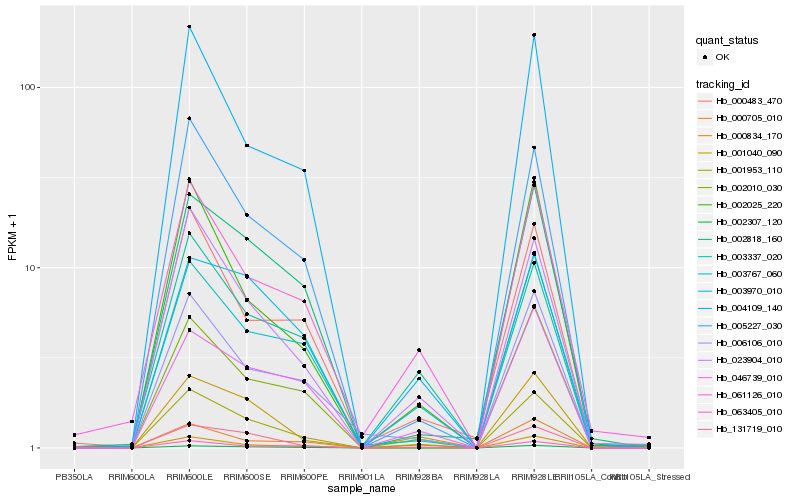
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001953_110 |
0.0 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 2 |
Hb_023904_010 |
0.083250435 |
- |
- |
- |
| 3 |
Hb_001040_090 |
0.095746288 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |
| 4 |
Hb_005227_030 |
0.0960253331 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002010_030 |
0.1037337164 |
- |
- |
Phytosulfokine receptor precursor, putative [Ricinus communis] |
| 6 |
Hb_004109_140 |
0.1097012896 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002818_160 |
0.1159089407 |
- |
- |
PREDICTED: uncharacterized protein LOC105642722 [Jatropha curcas] |
| 8 |
Hb_003767_060 |
0.118262311 |
- |
- |
PREDICTED: serine/threonine-protein kinase CDL1 [Jatropha curcas] |
| 9 |
Hb_000834_170 |
0.118344679 |
- |
- |
polygalacturonase, putative [Ricinus communis] |
| 10 |
Hb_002025_220 |
0.1184305408 |
transcription factor |
TF Family: C2H2 |
zinc finger protein, putative [Ricinus communis] |
| 11 |
Hb_006106_010 |
0.1202466972 |
transcription factor |
TF Family: TCP |
transcription factor, putative [Ricinus communis] |
| 12 |
Hb_131719_010 |
0.1203363792 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 13 |
Hb_002307_120 |
0.120625931 |
- |
- |
scab resistance family protein [Populus trichocarpa] |
| 14 |
Hb_000705_010 |
0.1215415174 |
- |
- |
hypothetical protein CICLE_v10027296mg [Citrus clementina] |
| 15 |
Hb_061126_010 |
0.1235482891 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 16 |
Hb_046739_010 |
0.1247271177 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g18390 isoform X1 [Jatropha curcas] |
| 17 |
Hb_003970_010 |
0.1254713782 |
- |
- |
PREDICTED: sigma factor binding protein 1, chloroplastic-like [Populus euphratica] |
| 18 |
Hb_063405_010 |
0.1272426926 |
- |
- |
PREDICTED: probable terpene synthase 3 [Jatropha curcas] |
| 19 |
Hb_000483_470 |
0.1286767576 |
- |
- |
PREDICTED: zeaxanthin epoxidase, chloroplastic-like isoform X1 [Jatropha curcas] |
| 20 |
Hb_003337_020 |
0.130046078 |
- |
- |
PREDICTED: uncharacterized protein LOC105116455 [Populus euphratica] |