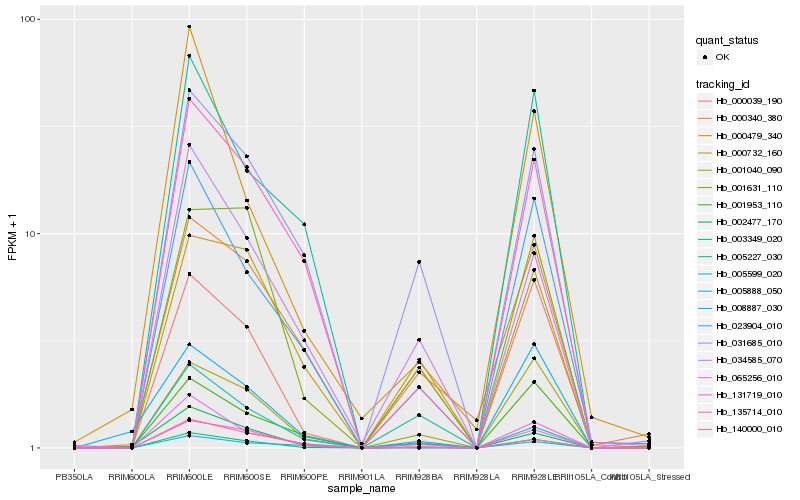
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003349_020 |
0.0 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 2 |
Hb_023904_010 |
0.1287764021 |
- |
- |
- |
| 3 |
Hb_001040_090 |
0.1305235804 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |
| 4 |
Hb_135714_010 |
0.1404313524 |
- |
- |
hypothetical protein POPTR_0017s09310g, partial [Populus trichocarpa] |
| 5 |
Hb_131719_010 |
0.1461254945 |
- |
- |
PREDICTED: uncharacterized protein LOC104107451 [Nicotiana tomentosiformis] |
| 6 |
Hb_001953_110 |
0.1467990529 |
- |
- |
PREDICTED: protein TAPETUM DETERMINANT 1-like [Jatropha curcas] |
| 7 |
Hb_031685_010 |
0.1493049746 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 8 |
Hb_001631_110 |
0.1525338621 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: gamma-glutamyltranspeptidase 1-like [Populus euphratica] |
| 9 |
Hb_034585_070 |
0.1532690205 |
- |
- |
Polyneuridine-aldehyde esterase precursor, putative [Ricinus communis] |
| 10 |
Hb_065256_010 |
0.1568141557 |
- |
- |
tryptophan synthase beta chain, putative [Ricinus communis] |
| 11 |
Hb_005888_050 |
0.1608830822 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 12 |
Hb_000039_190 |
0.1643774584 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 13 |
Hb_000732_160 |
0.1735052608 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 14 |
Hb_002477_170 |
0.1736690039 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 15 |
Hb_000340_380 |
0.1743928612 |
- |
- |
PREDICTED: receptor-like cytosolic serine/threonine-protein kinase RBK2 [Jatropha curcas] |
| 16 |
Hb_005227_030 |
0.1793955786 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP28, chloroplastic [Jatropha curcas] |
| 17 |
Hb_000479_340 |
0.1814899706 |
- |
- |
PREDICTED: uncharacterized protein LOC105644112 [Jatropha curcas] |
| 18 |
Hb_140000_010 |
0.1826183957 |
- |
- |
receptor-kinase, putative [Ricinus communis] |
| 19 |
Hb_008887_030 |
0.1852266693 |
- |
- |
PREDICTED: uncharacterized protein LOC105650448 [Jatropha curcas] |
| 20 |
Hb_005599_020 |
0.1862682239 |
- |
- |
Protein MLO, putative [Ricinus communis] |