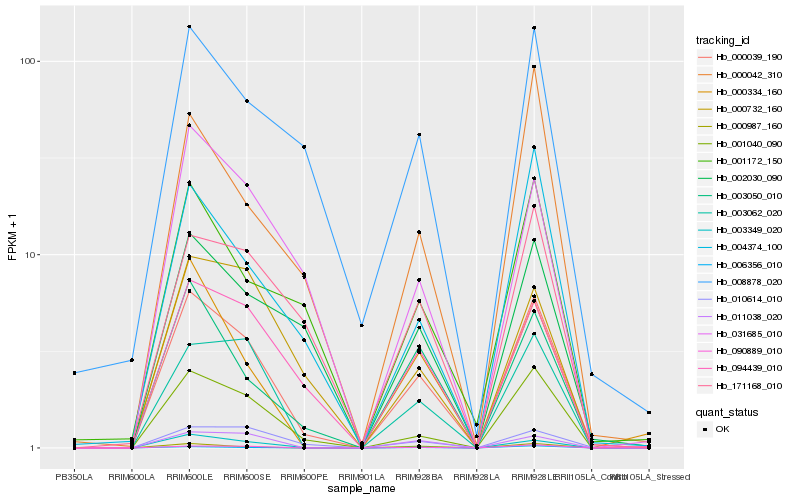
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000039_190 |
0.0 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 2 |
Hb_001040_090 |
0.1159354283 |
- |
- |
PREDICTED: tetraspanin-19-like [Jatropha curcas] |
| 3 |
Hb_090889_010 |
0.1440030097 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 4 |
Hb_094439_010 |
0.1501205537 |
- |
- |
PREDICTED: MACPF domain-containing protein At1g14780 [Jatropha curcas] |
| 5 |
Hb_003062_020 |
0.154017569 |
- |
- |
Polyprotein, putative [Solanum demissum] |
| 6 |
Hb_003050_010 |
0.1574042276 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 7 |
Hb_000987_160 |
0.1591306604 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 8 |
Hb_003349_020 |
0.1643774584 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 9 |
Hb_006356_010 |
0.1650819645 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 10 |
Hb_002030_090 |
0.1665082243 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like [Jatropha curcas] |
| 11 |
Hb_004374_100 |
0.1668876413 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |
| 12 |
Hb_011038_020 |
0.1675891943 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_010614_010 |
0.167929017 |
- |
- |
leucine-rich repeat transmembrane protein kinase [Populus trichocarpa] |
| 14 |
Hb_171168_010 |
0.1712542326 |
transcription factor |
TF Family: NAC |
NAC domain-containing protein, putative [Ricinus communis] |
| 15 |
Hb_031685_010 |
0.1741995046 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 16 |
Hb_008878_020 |
0.1758782531 |
- |
- |
PREDICTED: aldehyde dehydrogenase family 3 member H1-like [Populus euphratica] |
| 17 |
Hb_000334_160 |
0.1791008612 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_001172_150 |
0.1795329356 |
desease resistance |
Gene Name: ABC_membrane |
PREDICTED: putative ABC transporter C family member 15 [Jatropha curcas] |
| 19 |
Hb_000732_160 |
0.1808219603 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 20 |
Hb_000042_310 |
0.1815143159 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |