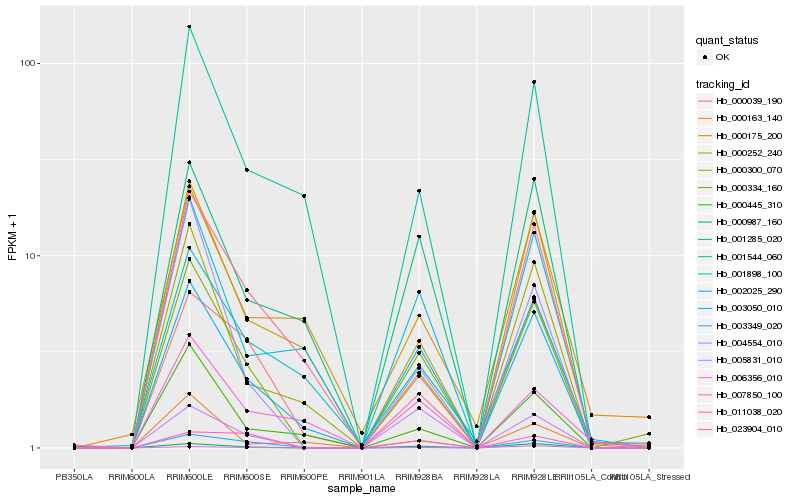
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000334_160 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000987_160 |
0.1342281488 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 3 |
Hb_003050_010 |
0.1346035875 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 4 |
Hb_000039_190 |
0.1791008612 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 5 |
Hb_004554_010 |
0.1853430671 |
- |
- |
PREDICTED: uncharacterized protein LOC105636233 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000300_070 |
0.1947613123 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 7 |
Hb_001285_020 |
0.1966229544 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 8 |
Hb_000445_310 |
0.2024950859 |
- |
- |
hypothetical protein B456_011G218000 [Gossypium raimondii] |
| 9 |
Hb_000252_240 |
0.2036398731 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X2 [Gossypium raimondii] |
| 10 |
Hb_002025_290 |
0.2039505753 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001544_060 |
0.2085685875 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription repressor MYB4 isoform X2 [Jatropha curcas] |
| 12 |
Hb_001898_100 |
0.2108002024 |
- |
- |
PREDICTED: phosphatidylserine decarboxylase proenzyme 2-like [Jatropha curcas] |
| 13 |
Hb_005831_010 |
0.2187439988 |
- |
- |
hypothetical protein JCGZ_04003 [Jatropha curcas] |
| 14 |
Hb_006356_010 |
0.2212620294 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 15 |
Hb_003349_020 |
0.2303613849 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: disease resistance protein At4g27190-like [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_000175_200 |
0.235418064 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_000163_140 |
0.2357112933 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 18 |
Hb_011038_020 |
0.2359663733 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_007850_100 |
0.2368161156 |
- |
- |
PREDICTED: uncharacterized protein LOC105640661 [Jatropha curcas] |
| 20 |
Hb_023904_010 |
0.2380333031 |
- |
- |
- |