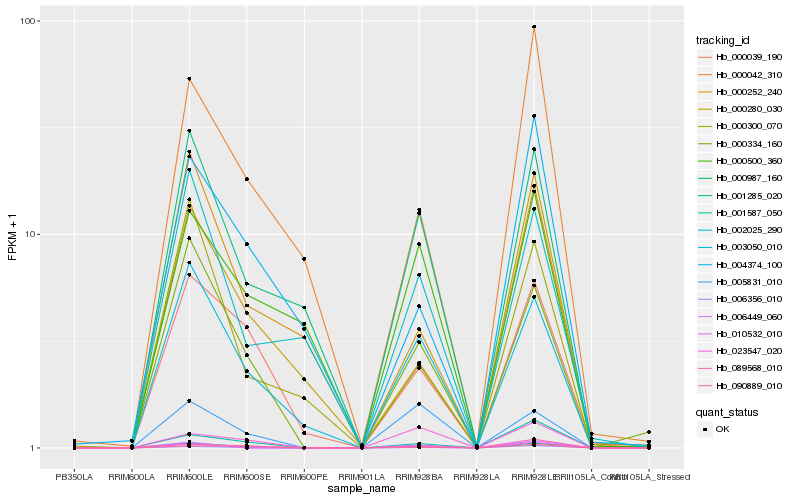
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000987_160 |
0.0 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 [Jatropha curcas] |
| 2 |
Hb_006356_010 |
0.1227452896 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 3 |
Hb_003050_010 |
0.1283386021 |
- |
- |
PREDICTED: VQ motif-containing protein 1-like [Jatropha curcas] |
| 4 |
Hb_000334_160 |
0.1342281488 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000039_190 |
0.1591306604 |
transcription factor |
TF Family: MYB |
PREDICTED: transcription factor MYB3-like [Jatropha curcas] |
| 6 |
Hb_001285_020 |
0.1640567687 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 7 |
Hb_089568_010 |
0.174555255 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 8 |
Hb_001587_050 |
0.1809108323 |
- |
- |
- |
| 9 |
Hb_005831_010 |
0.1817240613 |
- |
- |
hypothetical protein JCGZ_04003 [Jatropha curcas] |
| 10 |
Hb_002025_290 |
0.1990519933 |
transcription factor |
TF Family: G2-like |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_023547_020 |
0.2034436491 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_090889_010 |
0.2047770146 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Jatropha curcas] |
| 13 |
Hb_000042_310 |
0.2059467481 |
- |
- |
Cyclic nucleotide-gated ion channel, putative [Ricinus communis] |
| 14 |
Hb_000500_360 |
0.2060507917 |
- |
- |
Xylem serine proteinase 1 precursor, putative [Ricinus communis] |
| 15 |
Hb_000300_070 |
0.2098287392 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 16 |
Hb_000280_030 |
0.2128943675 |
transcription factor |
TF Family: NAC |
PREDICTED: NAC domain-containing protein 55-like [Jatropha curcas] |
| 17 |
Hb_010532_010 |
0.2137599957 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g08850 isoform X1 [Populus euphratica] |
| 18 |
Hb_006449_060 |
0.2139693952 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 19 |
Hb_004374_100 |
0.2155194455 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein AT74H [Jatropha curcas] |
| 20 |
Hb_000252_240 |
0.2156663115 |
- |
- |
PREDICTED: protein TRANSPARENT TESTA 12-like isoform X2 [Gossypium raimondii] |