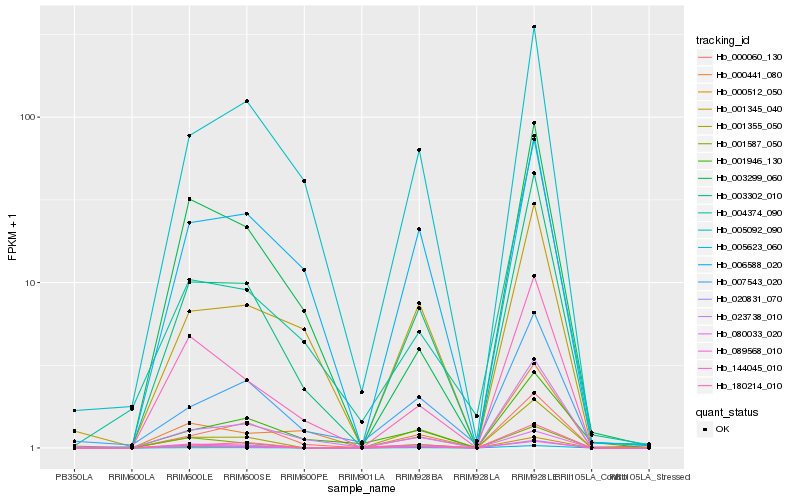
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003302_010 |
0.0 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 2 |
Hb_023738_010 |
0.1176608137 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 3 |
Hb_080033_020 |
0.1270390272 |
- |
- |
PREDICTED: probable leucine-rich repeat receptor-like protein kinase At1g35710 [Vitis vinifera] |
| 4 |
Hb_144045_010 |
0.1309178104 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription factor NGA1-like [Jatropha curcas] |
| 5 |
Hb_000512_050 |
0.1319150583 |
- |
- |
hypothetical protein VITISV_005640 [Vitis vinifera] |
| 6 |
Hb_005092_090 |
0.1392096627 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 7 |
Hb_000060_130 |
0.1402839909 |
- |
- |
- |
| 8 |
Hb_007543_020 |
0.1502036552 |
- |
- |
PREDICTED: uncharacterized protein LOC104229533, partial [Nicotiana sylvestris] |
| 9 |
Hb_089568_010 |
0.1512274635 |
- |
- |
ferric reductase oxidase [Manihot esculenta] |
| 10 |
Hb_005623_060 |
0.1534964409 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 11 |
Hb_004374_090 |
0.1546167501 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 12 |
Hb_001345_040 |
0.1546268083 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |
| 13 |
Hb_006588_020 |
0.155370009 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 14 |
Hb_000441_080 |
0.1580307539 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 15 |
Hb_003299_060 |
0.1596804056 |
- |
- |
nitrate transporter, putative [Ricinus communis] |
| 16 |
Hb_001587_050 |
0.1605167905 |
- |
- |
- |
| 17 |
Hb_001355_050 |
0.1622259882 |
- |
- |
- |
| 18 |
Hb_180214_010 |
0.1634289851 |
- |
- |
PREDICTED: glutamic acid-rich protein-like [Jatropha curcas] |
| 19 |
Hb_020831_070 |
0.164295509 |
- |
- |
hypothetical protein VITISV_027174 [Vitis vinifera] |
| 20 |
Hb_001946_130 |
0.1663466017 |
- |
- |
- |