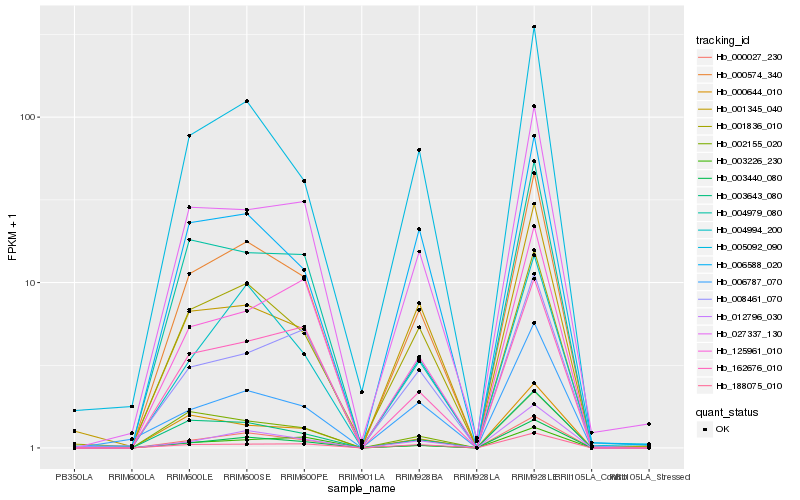
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000574_340 |
0.0 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 2 |
Hb_003440_080 |
0.0660869814 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 3 |
Hb_003643_080 |
0.0927113626 |
- |
- |
hypothetical protein JCGZ_01308 [Jatropha curcas] |
| 4 |
Hb_012796_030 |
0.0940926818 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 5 |
Hb_027337_130 |
0.0962599378 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 6 |
Hb_005092_090 |
0.0983095012 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 7 |
Hb_188075_010 |
0.0983614801 |
- |
- |
PREDICTED: uncharacterized protein LOC105775124 [Gossypium raimondii] |
| 8 |
Hb_006588_020 |
0.1145250382 |
transcription factor |
TF Family: G2-like |
DNA binding protein, putative [Ricinus communis] |
| 9 |
Hb_162676_010 |
0.1158429455 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 10 |
Hb_000644_010 |
0.1177785607 |
- |
- |
PREDICTED: putative disease resistance protein RGA1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000027_230 |
0.1211479234 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 12 |
Hb_004994_200 |
0.1211886524 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 13 |
Hb_004979_080 |
0.1222463489 |
- |
- |
kinase, putative [Ricinus communis] |
| 14 |
Hb_003226_230 |
0.1293508854 |
- |
- |
PREDICTED: transposon Ty3-G Gag-Pol polyprotein [Phoenix dactylifera] |
| 15 |
Hb_006787_070 |
0.1294635506 |
- |
- |
PREDICTED: uncharacterized protein LOC104121706, partial [Nicotiana tomentosiformis] |
| 16 |
Hb_001836_010 |
0.1297061221 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 17 |
Hb_125961_010 |
0.1323504026 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 18 |
Hb_002155_020 |
0.1333327437 |
- |
- |
PREDICTED: probable WRKY transcription factor 65 [Jatropha curcas] |
| 19 |
Hb_008461_070 |
0.1333378632 |
- |
- |
PREDICTED: ethylene-overproduction protein 1 isoform X1 [Jatropha curcas] |
| 20 |
Hb_001345_040 |
0.1336757456 |
- |
- |
hypothetical protein VITISV_015004 [Vitis vinifera] |