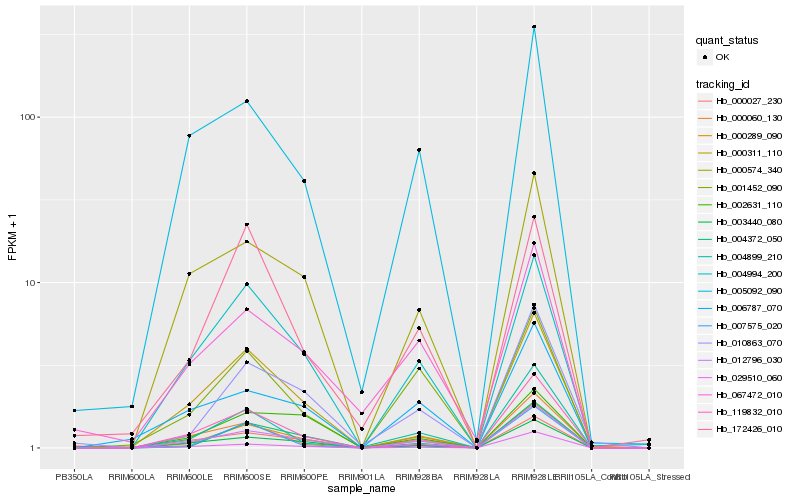
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000289_090 |
0.0 |
- |
- |
- |
| 2 |
Hb_012796_030 |
0.0705170054 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 3 |
Hb_003440_080 |
0.1080904621 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 4 |
Hb_004994_200 |
0.1120854395 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 5 |
Hb_010863_070 |
0.1186530091 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 6 |
Hb_119832_010 |
0.1267743793 |
- |
- |
- |
| 7 |
Hb_004372_050 |
0.1359358511 |
- |
- |
putative receptor-like protein kinase [Morus notabilis] |
| 8 |
Hb_000311_110 |
0.1365613138 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 9 |
Hb_007575_020 |
0.139671954 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 10 |
Hb_000574_340 |
0.1409173741 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 11 |
Hb_004899_210 |
0.1478162534 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein E [Jatropha curcas] |
| 12 |
Hb_005092_090 |
0.1520706939 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 13 |
Hb_029510_060 |
0.1525042712 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 14 |
Hb_001452_090 |
0.1548758863 |
- |
- |
PREDICTED: serine carboxypeptidase II-3-like [Vitis vinifera] |
| 15 |
Hb_000027_230 |
0.1637072485 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 16 |
Hb_006787_070 |
0.1647634938 |
- |
- |
PREDICTED: uncharacterized protein LOC104121706, partial [Nicotiana tomentosiformis] |
| 17 |
Hb_002631_110 |
0.1668368135 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 18 |
Hb_172426_010 |
0.1670483236 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 19 |
Hb_000060_130 |
0.1676466509 |
- |
- |
- |
| 20 |
Hb_067472_010 |
0.1702677133 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |