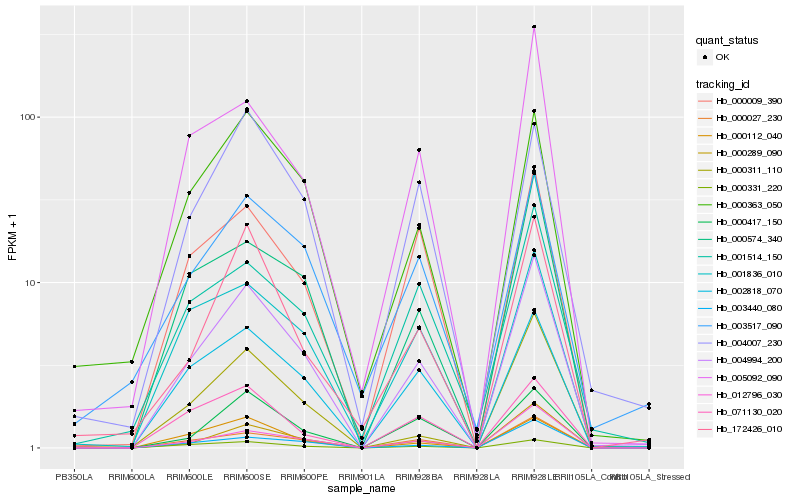
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004994_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |
| 2 |
Hb_000289_090 |
0.1120854395 |
- |
- |
- |
| 3 |
Hb_000331_220 |
0.1151681711 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_002818_070 |
0.1165496584 |
transcription factor |
TF Family: WRKY |
WRKY transcription factor, putative [Ricinus communis] |
| 5 |
Hb_000574_340 |
0.1211886524 |
- |
- |
hypothetical protein RCOM_0901420 [Ricinus communis] |
| 6 |
Hb_012796_030 |
0.1258162532 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 7 |
Hb_001836_010 |
0.132728679 |
- |
- |
PREDICTED: G-type lectin S-receptor-like serine/threonine-protein kinase At4g03230 [Jatropha curcas] |
| 8 |
Hb_003440_080 |
0.1347448531 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 9 |
Hb_005092_090 |
0.1353106314 |
- |
- |
cold regulated protein [Manihot esculenta] |
| 10 |
Hb_000027_230 |
0.135349129 |
- |
- |
PREDICTED: probable receptor-like protein kinase At1g67000 [Jatropha curcas] |
| 11 |
Hb_000112_040 |
0.1376507164 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 12 |
Hb_172426_010 |
0.1377326908 |
- |
- |
PREDICTED: heavy metal-associated isoprenylated plant protein 26 [Jatropha curcas] |
| 13 |
Hb_000311_110 |
0.1378850436 |
- |
- |
ATP-binding cassette transporter, putative [Ricinus communis] |
| 14 |
Hb_000417_150 |
0.1381897155 |
- |
- |
hypothetical protein JCGZ_21586 [Jatropha curcas] |
| 15 |
Hb_000363_050 |
0.1391018948 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 16 |
Hb_000009_390 |
0.1404534972 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_003517_090 |
0.1459056933 |
- |
- |
PREDICTED: zinc finger CCCH domain-containing protein 18-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_001514_150 |
0.1467709735 |
- |
- |
PREDICTED: cytochrome P450 81E8-like [Jatropha curcas] |
| 19 |
Hb_004007_230 |
0.1487411882 |
- |
- |
PREDICTED: putative formamidase C869.04 isoform X1 [Jatropha curcas] |
| 20 |
Hb_071130_020 |
0.1487545292 |
- |
- |
- |