| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
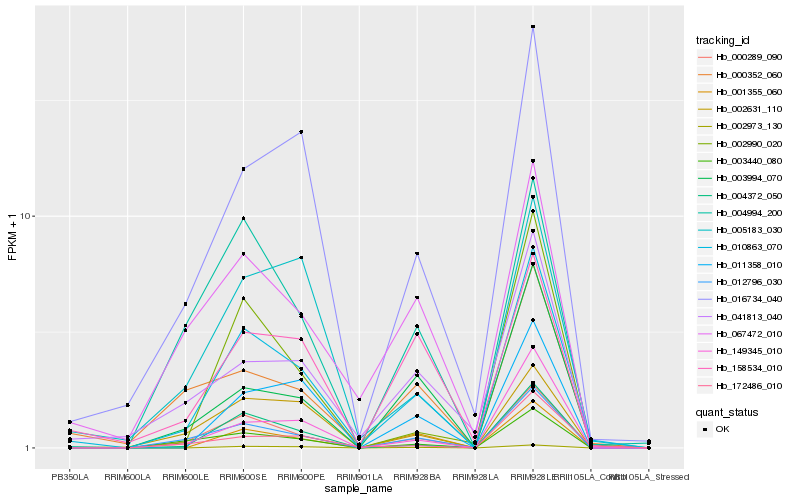
Hb_010863_070 |
0.0 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 2 |
Hb_000289_090 |
0.1186530091 |
- |
- |
- |
| 3 |
Hb_012796_030 |
0.137770677 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 4 |
Hb_004372_050 |
0.1466574233 |
- |
- |
putative receptor-like protein kinase [Morus notabilis] |
| 5 |
Hb_016734_040 |
0.1558308772 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 6 |
Hb_149345_010 |
0.1615423613 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 7 |
Hb_003440_080 |
0.1674499666 |
- |
- |
polyprotein [Citrus endogenous pararetrovirus] |
| 8 |
Hb_001355_060 |
0.1720467981 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 9 |
Hb_003994_070 |
0.1753367543 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 10 |
Hb_067472_010 |
0.1765724315 |
- |
- |
Uncharacterized protein TCM_024268 [Theobroma cacao] |
| 11 |
Hb_002973_130 |
0.1773072652 |
- |
- |
PREDICTED: uncharacterized protein LOC104609908 [Nelumbo nucifera] |
| 12 |
Hb_005183_030 |
0.1785291498 |
- |
- |
PREDICTED: probable calcium-binding protein CML16 [Jatropha curcas] |
| 13 |
Hb_002631_110 |
0.1786854554 |
- |
- |
PREDICTED: cytochrome P450 71D9-like [Fragaria vesca subsp. vesca] |
| 14 |
Hb_011358_010 |
0.1794508912 |
- |
- |
- |
| 15 |
Hb_041813_040 |
0.1801184893 |
- |
- |
NADH2 dehydrogenase (ubiquinone) (EC 1.6.5.3) chain 5, truncated - sugar beet mitochondrion (mitochondrion) [Beta vulgaris subsp. vulgaris] |
| 16 |
Hb_000352_060 |
0.1802748527 |
- |
- |
RNA-directed DNA polymerase [Arachis hypogaea] |
| 17 |
Hb_002990_020 |
0.1812969059 |
- |
- |
PREDICTED: vacuolar amino acid transporter 1 [Jatropha curcas] |
| 18 |
Hb_172486_010 |
0.1813182416 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 19 |
Hb_158534_010 |
0.1822792101 |
desease resistance |
Gene Name: NB-ARC |
NBS-LRR resistance protein RGH1 [Manihot esculenta] |
| 20 |
Hb_004994_200 |
0.1838019871 |
- |
- |
PREDICTED: uncharacterized protein LOC105174340 [Sesamum indicum] |