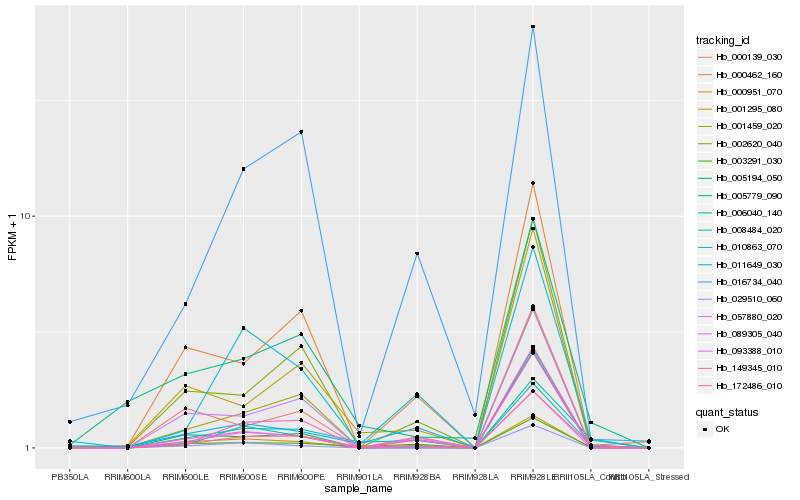
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_172486_010 |
0.0 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 2 |
Hb_008484_020 |
0.1391755735 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 3 |
Hb_149345_010 |
0.1400473369 |
- |
- |
Anthranilate N-benzoyltransferase protein, putative [Ricinus communis] |
| 4 |
Hb_002620_040 |
0.1520603209 |
- |
- |
PREDICTED: calmodulin-binding receptor-like cytoplasmic kinase 2 [Jatropha curcas] |
| 5 |
Hb_001459_020 |
0.152975965 |
- |
- |
ankyrin repeat protein [Bruguiera gymnorhiza] |
| 6 |
Hb_003291_030 |
0.1598048921 |
- |
- |
Gag-pol protein, putative [Solanum demissum] |
| 7 |
Hb_029510_060 |
0.1635802245 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 8 |
Hb_057880_020 |
0.1649593318 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1, partial [Jatropha curcas] |
| 9 |
Hb_001295_080 |
0.1669669334 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 10 |
Hb_093388_010 |
0.1669823574 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 11 |
Hb_000462_160 |
0.1724067504 |
- |
- |
UDP-glucosyltransferase, putative [Ricinus communis] |
| 12 |
Hb_006040_140 |
0.1730709761 |
- |
- |
hypothetical protein NitaMp105 [Nicotiana tabacum] |
| 13 |
Hb_089305_040 |
0.1736085097 |
- |
- |
- |
| 14 |
Hb_000139_030 |
0.1784162217 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 15 |
Hb_010863_070 |
0.1813182416 |
- |
- |
PREDICTED: glutaredoxin-C11 [Jatropha curcas] |
| 16 |
Hb_000951_070 |
0.181815668 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: receptor-like protein kinase FERONIA [Populus euphratica] |
| 17 |
Hb_005194_050 |
0.1843324872 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 18 |
Hb_011649_030 |
0.1857027497 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 19 |
Hb_016734_040 |
0.1876537669 |
- |
- |
abhydrolase domain containing, putative [Ricinus communis] |
| 20 |
Hb_005779_090 |
0.189106599 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |