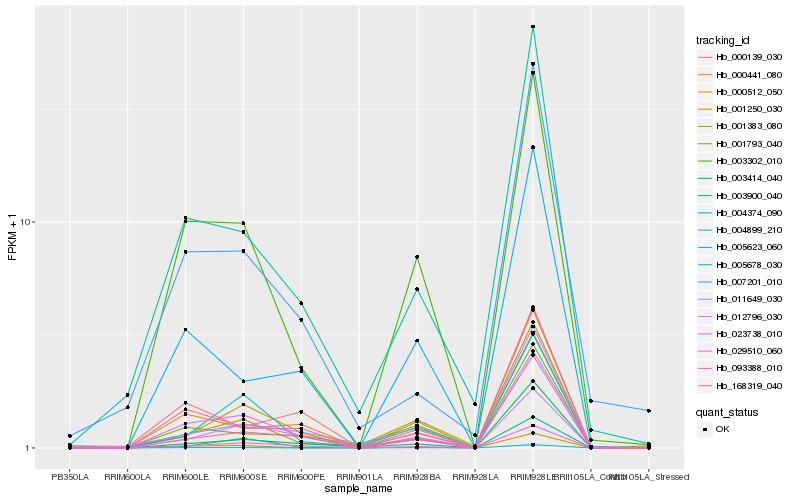
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_023738_010 |
0.0 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 2 |
Hb_029510_060 |
0.091164889 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 3 |
Hb_093388_010 |
0.0974368679 |
- |
- |
PREDICTED: LRR receptor-like serine/threonine-protein kinase GSO1 [Jatropha curcas] |
| 4 |
Hb_004374_090 |
0.1044322741 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 5 |
Hb_001250_030 |
0.1149435567 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003302_010 |
0.1176608137 |
- |
- |
PREDICTED: alpha-dioxygenase 1 [Jatropha curcas] |
| 7 |
Hb_000441_080 |
0.1291510648 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 8 |
Hb_168319_040 |
0.1343553468 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 9 |
Hb_003414_040 |
0.1344127892 |
- |
- |
hypothetical protein VITISV_016691 [Vitis vinifera] |
| 10 |
Hb_000512_050 |
0.1420638419 |
- |
- |
hypothetical protein VITISV_005640 [Vitis vinifera] |
| 11 |
Hb_001383_080 |
0.1437368905 |
- |
- |
OSJNBa0019D11.8 [Oryza sativa Japonica Group] |
| 12 |
Hb_005678_030 |
0.1440400996 |
- |
- |
PREDICTED: uncharacterized protein At4g06744-like [Jatropha curcas] |
| 13 |
Hb_004899_210 |
0.1511408793 |
- |
- |
PREDICTED: respiratory burst oxidase homolog protein E [Jatropha curcas] |
| 14 |
Hb_007201_010 |
0.1536800917 |
- |
- |
PREDICTED: uncharacterized protein LOC105631226 isoform X2 [Jatropha curcas] |
| 15 |
Hb_001793_040 |
0.1561852744 |
- |
- |
hypothetical protein VITISV_006810 [Vitis vinifera] |
| 16 |
Hb_011649_030 |
0.1562548021 |
- |
- |
hypothetical protein [Vitis hybrid cultivar] |
| 17 |
Hb_005623_060 |
0.1564070259 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_012796_030 |
0.1567085426 |
- |
- |
PREDICTED: uncharacterized protein LOC104106381 [Nicotiana tomentosiformis] |
| 19 |
Hb_000139_030 |
0.1571540704 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 20 |
Hb_003900_040 |
0.1593209536 |
- |
- |
hypothetical protein [Lotus japonicus] |