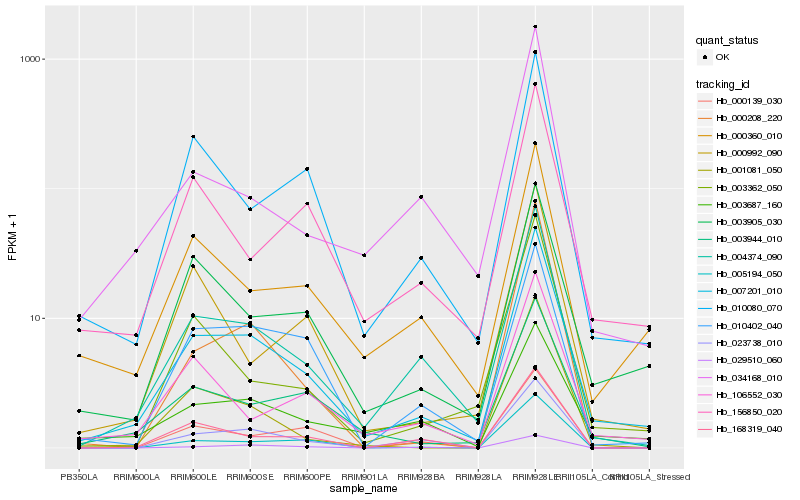
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007201_010 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105631226 isoform X2 [Jatropha curcas] |
| 2 |
Hb_004374_090 |
0.1122130226 |
transcription factor |
TF Family: HB |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 3 |
Hb_003944_010 |
0.1348887109 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 4 |
Hb_003905_030 |
0.1395093738 |
- |
- |
PREDICTED: clustered mitochondria protein homolog [Jatropha curcas] |
| 5 |
Hb_000360_010 |
0.1456825068 |
- |
- |
PREDICTED: ferredoxin-dependent glutamate synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 6 |
Hb_010080_070 |
0.1509736576 |
- |
- |
PREDICTED: ATP-dependent zinc metalloprotease FTSH, chloroplastic [Jatropha curcas] |
| 7 |
Hb_001081_050 |
0.1529273595 |
- |
- |
hypothetical protein POPTR_0001s07860g [Populus trichocarpa] |
| 8 |
Hb_023738_010 |
0.1536800917 |
- |
- |
Concanavalin A-like lectin protein kinase family protein [Theobroma cacao] |
| 9 |
Hb_106552_030 |
0.1561857974 |
- |
- |
hypothetical protein POPTR_0006s23180g [Populus trichocarpa] |
| 10 |
Hb_003687_160 |
0.1570841933 |
- |
- |
Ran GTPase binding protein, putative [Ricinus communis] |
| 11 |
Hb_156850_020 |
0.1580755487 |
- |
- |
ATP-dependent zinc metalloprotease FTSH 1, chloroplastic [Glycine soja] |
| 12 |
Hb_010402_040 |
0.1584714704 |
- |
- |
hypothetical protein JCGZ_21227 [Jatropha curcas] |
| 13 |
Hb_168319_040 |
0.1603298541 |
- |
- |
PREDICTED: cytochrome P450 CYP749A22-like [Jatropha curcas] |
| 14 |
Hb_003362_050 |
0.1603332694 |
- |
- |
PREDICTED: probable sucrose-phosphate synthase 3 [Jatropha curcas] |
| 15 |
Hb_005194_050 |
0.1613872161 |
transcription factor |
TF Family: GRAS |
SCL domain class transcription factor [Theobroma cacao] |
| 16 |
Hb_029510_060 |
0.1653086715 |
- |
- |
PREDICTED: probable plastidic glucose transporter 3 [Jatropha curcas] |
| 17 |
Hb_034168_010 |
0.1695656722 |
- |
- |
hypothetical protein [Azospirillum sp. CAG:260] |
| 18 |
Hb_000139_030 |
0.1697775591 |
- |
- |
PREDICTED: uncharacterized protein LOC101254423 [Solanum lycopersicum] |
| 19 |
Hb_000992_090 |
0.1704281322 |
transcription factor |
TF Family: Orphans |
CIL, putative [Ricinus communis] |
| 20 |
Hb_000208_220 |
0.1710631274 |
- |
- |
- |