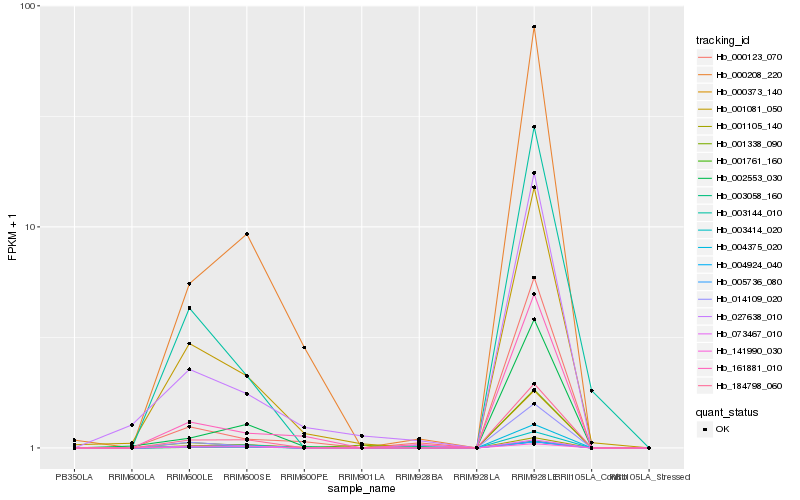
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_184798_060 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001761_160 |
0.0903636319 |
- |
- |
hypothetical protein VITISV_019364 [Vitis vinifera] |
| 3 |
Hb_000208_220 |
0.1151640443 |
- |
- |
- |
| 4 |
Hb_001081_050 |
0.1155532092 |
- |
- |
hypothetical protein POPTR_0001s07860g [Populus trichocarpa] |
| 5 |
Hb_002553_030 |
0.1186228079 |
- |
- |
hypothetical protein POPTR_0008s16550g [Populus trichocarpa] |
| 6 |
Hb_003414_020 |
0.126257429 |
- |
- |
PREDICTED: uncharacterized protein LOC103655285 [Zea mays] |
| 7 |
Hb_003144_010 |
0.142217566 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 8 |
Hb_004924_040 |
0.1459811053 |
- |
- |
PREDICTED: uncharacterized protein LOC105650390, partial [Jatropha curcas] |
| 9 |
Hb_003058_160 |
0.1463275699 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_073467_010 |
0.1474735082 |
- |
- |
PREDICTED: uncharacterized protein LOC105634453 [Jatropha curcas] |
| 11 |
Hb_027638_010 |
0.154508736 |
- |
- |
Protein KES1, putative [Ricinus communis] |
| 12 |
Hb_161881_010 |
0.1551169528 |
- |
- |
laccase, putative [Ricinus communis] |
| 13 |
Hb_001338_090 |
0.1609881186 |
- |
- |
hypothetical protein VITISV_009275 [Vitis vinifera] |
| 14 |
Hb_141990_030 |
0.1642172788 |
- |
- |
hypothetical protein JCGZ_09181 [Jatropha curcas] |
| 15 |
Hb_005736_080 |
0.1679745015 |
- |
- |
hypothetical protein CICLE_v10033628mg, partial [Citrus clementina] |
| 16 |
Hb_000373_140 |
0.1713242894 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 17 |
Hb_014109_020 |
0.1722194078 |
transcription factor |
TF Family: WRKY |
PREDICTED: probable WRKY transcription factor 53 [Jatropha curcas] |
| 18 |
Hb_000123_070 |
0.173315157 |
- |
- |
PREDICTED: terpene synthase 10-like [Jatropha curcas] |
| 19 |
Hb_001105_140 |
0.1751190473 |
- |
- |
Alpha-expansin 1 precursor, putative [Ricinus communis] |
| 20 |
Hb_004375_020 |
0.1759627488 |
- |
- |
hypothetical protein VITISV_034560 [Vitis vinifera] |