| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
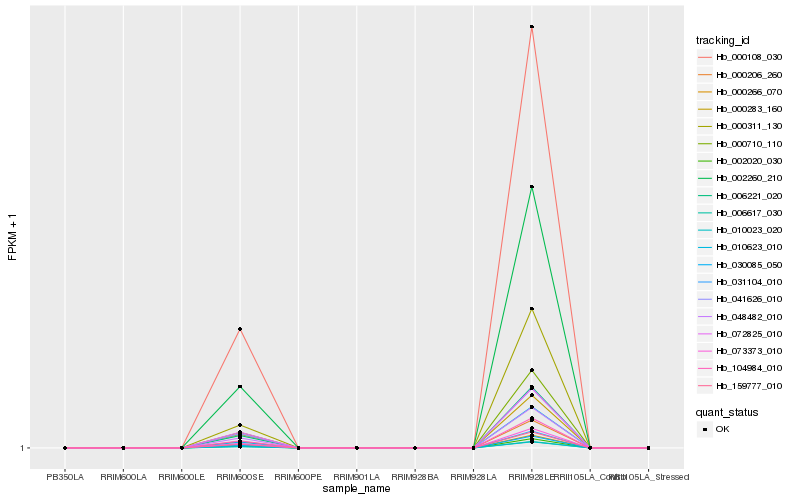
Hb_006221_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC104227769, partial [Nicotiana sylvestris] |
| 2 |
Hb_000710_110 |
0.0055232909 |
- |
- |
Vps51/Vps67 [Citrus endogenous pararetrovirus] |
| 3 |
Hb_000108_030 |
0.0141652372 |
- |
- |
- |
| 4 |
Hb_002260_210 |
0.0141937367 |
- |
- |
unnamed protein product [Vitis vinifera] |
| 5 |
Hb_010023_020 |
0.020879617 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 6 |
Hb_006617_030 |
0.0210153602 |
- |
- |
hypothetical protein PRUPE_ppa015000mg [Prunus persica] |
| 7 |
Hb_002020_030 |
0.0220675291 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 8 |
Hb_030085_050 |
0.0230566249 |
- |
- |
PREDICTED: uncharacterized protein LOC105797448 [Gossypium raimondii] |
| 9 |
Hb_000266_070 |
0.023377348 |
desease resistance |
Gene Name: NB-ARC |
PREDICTED: uncharacterized protein LOC105765578 [Gossypium raimondii] |
| 10 |
Hb_159777_010 |
0.0246494215 |
- |
- |
JHL22C18.10 [Jatropha curcas] |
| 11 |
Hb_073373_010 |
0.0260690965 |
- |
- |
hypothetical protein JCGZ_11284 [Jatropha curcas] |
| 12 |
Hb_041626_010 |
0.0266450169 |
- |
- |
PREDICTED: uncharacterized protein LOC105631130 [Jatropha curcas] |
| 13 |
Hb_000206_260 |
0.0294580053 |
- |
- |
cysteine protease, putative [Ricinus communis] |
| 14 |
Hb_104984_010 |
0.0303459407 |
- |
- |
PREDICTED: uncharacterized protein LOC105650298, partial [Jatropha curcas] |
| 15 |
Hb_048482_010 |
0.0350442128 |
- |
- |
- |
| 16 |
Hb_031104_010 |
0.0375895008 |
- |
- |
PREDICTED: uncharacterized protein LOC105800880, partial [Gossypium raimondii] |
| 17 |
Hb_000283_160 |
0.0407900197 |
- |
- |
hypothetical protein JCGZ_23510 [Jatropha curcas] |
| 18 |
Hb_072825_010 |
0.0442490196 |
- |
- |
Uncharacterized protein TCM_023328 [Theobroma cacao] |
| 19 |
Hb_000311_130 |
0.0460694804 |
- |
- |
PREDICTED: uncharacterized protein LOC105160092 [Sesamum indicum] |
| 20 |
Hb_010623_010 |
0.0497820196 |
- |
- |
PREDICTED: uncharacterized protein LOC105643732 [Jatropha curcas] |