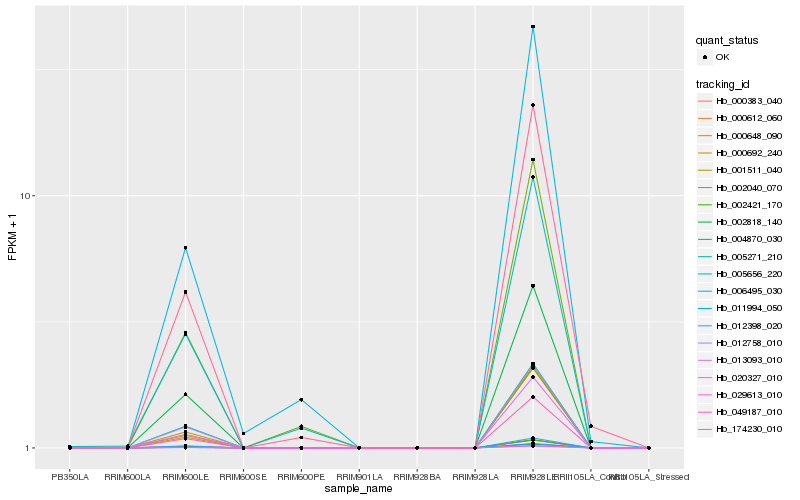
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_174230_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_19971 [Jatropha curcas] |
| 2 |
Hb_006495_030 |
0.0764251551 |
- |
- |
PREDICTED: isoflavone 2'-hydroxylase-like [Vitis vinifera] |
| 3 |
Hb_020327_010 |
0.0794869449 |
- |
- |
PREDICTED: uncharacterized protein LOC104886355, partial [Beta vulgaris subsp. vulgaris] |
| 4 |
Hb_000692_240 |
0.0795629993 |
- |
- |
Plastocyanin A, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_049187_010 |
0.0796415315 |
- |
- |
hypothetical protein, partial [Pseudomonas fluorescens] |
| 6 |
Hb_002040_070 |
0.08130402 |
- |
- |
- |
| 7 |
Hb_001511_040 |
0.085132893 |
- |
- |
PREDICTED: pistil-specific extensin-like protein [Jatropha curcas] |
| 8 |
Hb_012758_010 |
0.0877779994 |
- |
- |
PREDICTED: flowering-promoting factor 1-like protein 2 [Jatropha curcas] |
| 9 |
Hb_002818_140 |
0.0878081389 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_005271_210 |
0.0892348114 |
- |
- |
hypothetical protein JCGZ_20628 [Jatropha curcas] |
| 11 |
Hb_002421_170 |
0.0906517502 |
- |
- |
hypothetical protein POPTR_0003s18240g [Populus trichocarpa] |
| 12 |
Hb_011994_050 |
0.0920049624 |
- |
- |
multicopper oxidase, putative [Ricinus communis] |
| 13 |
Hb_004870_030 |
0.0926281238 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 14 |
Hb_005656_220 |
0.0926543418 |
- |
- |
PREDICTED: uncharacterized protein LOC105109832 [Populus euphratica] |
| 15 |
Hb_000612_060 |
0.094099519 |
- |
- |
PREDICTED: uncharacterized protein LOC105634770 [Jatropha curcas] |
| 16 |
Hb_000383_040 |
0.0941636755 |
- |
- |
PREDICTED: LOW QUALITY PROTEIN: uncharacterized protein LOC103696607 [Phoenix dactylifera] |
| 17 |
Hb_029613_010 |
0.094712394 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 18 |
Hb_013093_010 |
0.0950297863 |
- |
- |
retrotransposon protein, putative, Ty1-copia subclass [Oryza sativa Japonica Group] |
| 19 |
Hb_000648_090 |
0.0951893791 |
- |
- |
- |
| 20 |
Hb_012398_020 |
0.0954965912 |
- |
- |
hypothetical protein RCOM_0641040 [Ricinus communis] |