| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
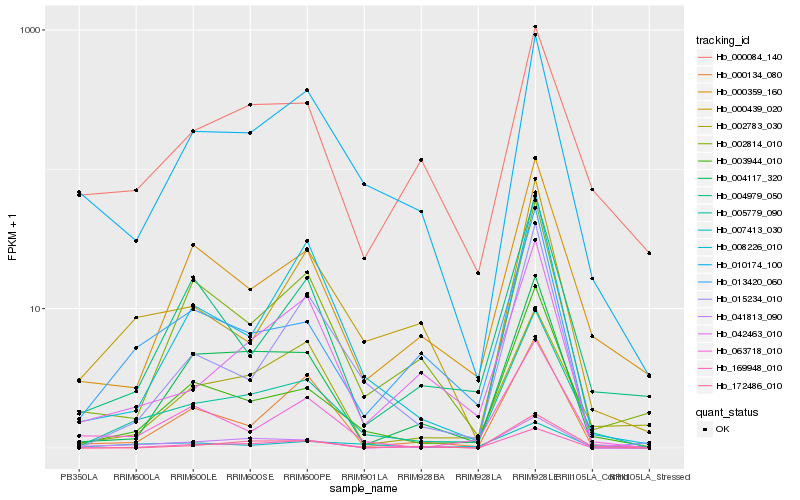
Hb_005779_090 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 2 |
Hb_003944_010 |
0.1348182369 |
- |
- |
PREDICTED: receptor-like protein kinase HSL1 [Jatropha curcas] |
| 3 |
Hb_000359_160 |
0.1754475163 |
- |
- |
PREDICTED: protein CAJ1 [Jatropha curcas] |
| 4 |
Hb_063718_010 |
0.1763003573 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_007413_030 |
0.1777631972 |
- |
- |
kinase, putative [Ricinus communis] |
| 6 |
Hb_013420_060 |
0.181969253 |
- |
- |
NADH dehydrogenase subunit 7 [Nymphaea sp. Bergthorsson 0401] |
| 7 |
Hb_004117_320 |
0.1822201005 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase WNK5 [Jatropha curcas] |
| 8 |
Hb_041813_090 |
0.1864014891 |
- |
- |
hypothetical protein EUGRSUZ_J01097 [Eucalyptus grandis] |
| 9 |
Hb_172486_010 |
0.189106599 |
- |
- |
leucine-rich repeat receptor protein kinase exs precursor, putative [Ricinus communis] |
| 10 |
Hb_008226_010 |
0.1900361198 |
- |
- |
PREDICTED: uncharacterized protein LOC105649179 [Jatropha curcas] |
| 11 |
Hb_010174_100 |
0.1911037544 |
- |
- |
PREDICTED: photosystem II core complex proteins psbY, chloroplastic-like [Populus euphratica] |
| 12 |
Hb_042463_010 |
0.1915159002 |
- |
- |
cytochrome c oxidase subunit 2-1 (mitochondrion) (mitochondrion) [Brassica juncea var. tumida] |
| 13 |
Hb_000439_020 |
0.1931658653 |
transcription factor |
TF Family: ERF |
PREDICTED: dehydration-responsive element-binding protein 3-like [Jatropha curcas] |
| 14 |
Hb_015234_010 |
0.1934959681 |
- |
- |
PREDICTED: oxysterol-binding protein-related protein 4B [Jatropha curcas] |
| 15 |
Hb_004979_050 |
0.1947014609 |
- |
- |
hypothetical protein JCGZ_23771 [Jatropha curcas] |
| 16 |
Hb_000084_140 |
0.195094968 |
- |
- |
Aspartic proteinase nepenthesin-2 precursor, putative [Ricinus communis] |
| 17 |
Hb_002814_010 |
0.1953315748 |
- |
- |
PREDICTED: adenylate kinase 5, chloroplastic [Jatropha curcas] |
| 18 |
Hb_002783_030 |
0.1970095703 |
- |
- |
PREDICTED: receptor protein kinase CLAVATA1 [Jatropha curcas] |
| 19 |
Hb_000134_080 |
0.1974476018 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 20 |
Hb_169948_010 |
0.1979992161 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |