| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
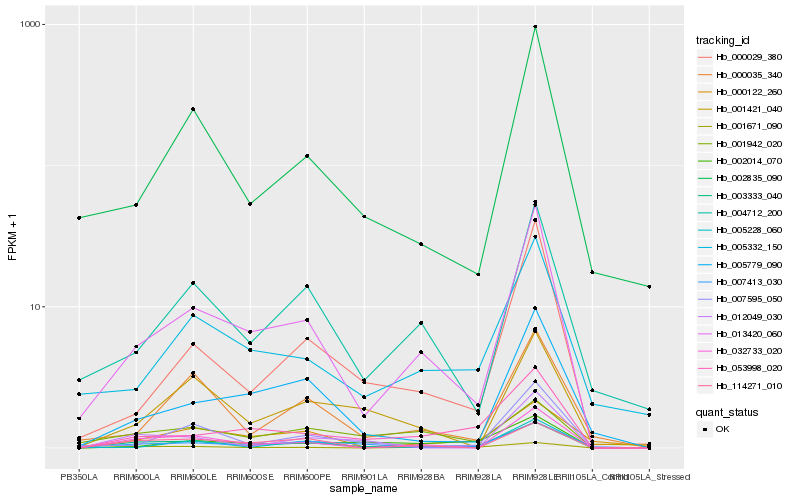
Hb_003333_040 |
0.0 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 2 |
Hb_013420_060 |
0.1652833146 |
- |
- |
NADH dehydrogenase subunit 7 [Nymphaea sp. Bergthorsson 0401] |
| 3 |
Hb_001671_090 |
0.1697958903 |
- |
- |
DNA/RNA polymerases superfamily protein [Theobroma cacao] |
| 4 |
Hb_007413_030 |
0.1891479009 |
- |
- |
kinase, putative [Ricinus communis] |
| 5 |
Hb_032733_020 |
0.1953555837 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X4 [Citrus sinensis] |
| 6 |
Hb_053998_020 |
0.2007017985 |
- |
- |
- |
| 7 |
Hb_114271_010 |
0.2120533589 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 8 |
Hb_000122_260 |
0.2219947593 |
transcription factor |
TF Family: SNF2 |
PREDICTED: SNF2 domain-containing protein CLASSY 1-like [Jatropha curcas] |
| 9 |
Hb_002835_090 |
0.2224990275 |
- |
- |
RecName: Full=Elongation factor Tu, chloroplastic; Short=EF-Tu; Flags: Precursor [Glycine max] |
| 10 |
Hb_000029_380 |
0.2261448196 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_012049_030 |
0.2279108827 |
- |
- |
RNA polymerase beta subunit [Hevea brasiliensis] |
| 12 |
Hb_005779_090 |
0.2291361064 |
transcription factor |
TF Family: ERF |
hypothetical protein POPTR_0014s05500g [Populus trichocarpa] |
| 13 |
Hb_002014_070 |
0.2306309796 |
- |
- |
PREDICTED: probable chlorophyll(ide) b reductase NYC1, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_007595_050 |
0.2310339 |
- |
- |
hypothetical chloroplast RF2 [Hevea brasiliensis] |
| 15 |
Hb_005228_060 |
0.2310897097 |
- |
- |
PREDICTED: uncharacterized protein LOC102604101 [Solanum tuberosum] |
| 16 |
Hb_005332_150 |
0.2331642683 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At3g46610 [Jatropha curcas] |
| 17 |
Hb_004712_200 |
0.2336711869 |
- |
- |
hypothetical protein CICLE_v10010292mg [Citrus clementina] |
| 18 |
Hb_001942_020 |
0.2366418433 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase FKBP16-3, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000035_340 |
0.2379990609 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001421_040 |
0.2396028532 |
- |
- |
PREDICTED: type I inositol 1,4,5-trisphosphate 5-phosphatase 2 [Jatropha curcas] |