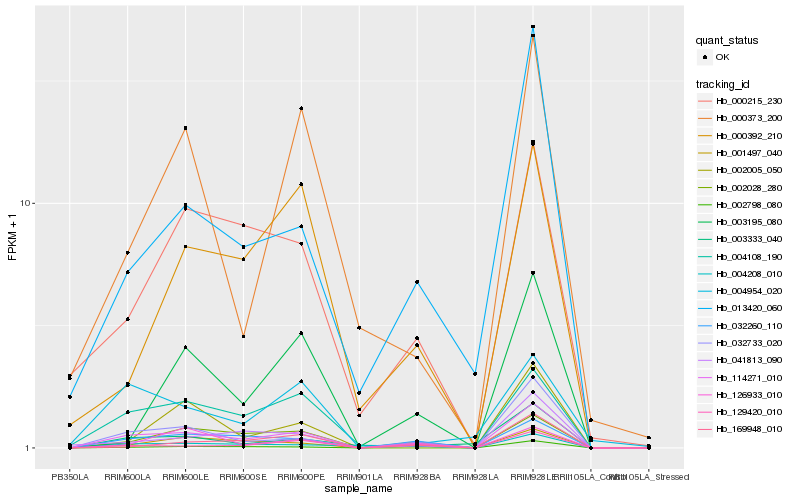
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_114271_010 |
0.0 |
- |
- |
hypothetical protein JCGZ_19461 [Jatropha curcas] |
| 2 |
Hb_032260_110 |
0.1498843769 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_032733_020 |
0.1583944559 |
- |
- |
PREDICTED: probable LRR receptor-like serine/threonine-protein kinase At4g36180-like isoform X4 [Citrus sinensis] |
| 4 |
Hb_004108_190 |
0.1755528806 |
- |
- |
PREDICTED: protein catecholamines up-like [Jatropha curcas] |
| 5 |
Hb_129420_010 |
0.1929397857 |
transcription factor |
TF Family: NAC |
NAC transcription factor 059 [Jatropha curcas] |
| 6 |
Hb_013420_060 |
0.1989542672 |
- |
- |
NADH dehydrogenase subunit 7 [Nymphaea sp. Bergthorsson 0401] |
| 7 |
Hb_002005_050 |
0.199870888 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 8 |
Hb_041813_090 |
0.2053149156 |
- |
- |
hypothetical protein EUGRSUZ_J01097 [Eucalyptus grandis] |
| 9 |
Hb_000215_230 |
0.2076841084 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 10 |
Hb_001497_040 |
0.2108882166 |
- |
- |
hypothetical protein POPTR_0008s219701g, partial [Populus trichocarpa] |
| 11 |
Hb_002028_280 |
0.2115257772 |
- |
- |
hypothetical protein RCOM_1397400 [Ricinus communis] |
| 12 |
Hb_003333_040 |
0.2120533589 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_169948_010 |
0.2133016439 |
- |
- |
hypothetical protein VITISV_006524 [Vitis vinifera] |
| 14 |
Hb_004208_010 |
0.2156740968 |
- |
- |
PREDICTED: uncharacterized protein LOC105914588 [Setaria italica] |
| 15 |
Hb_002798_080 |
0.2191071875 |
- |
- |
hypothetical protein VITISV_043745 [Vitis vinifera] |
| 16 |
Hb_000373_200 |
0.2294986677 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 17 |
Hb_003195_080 |
0.2302468599 |
- |
- |
PREDICTED: cytochrome P450 87A3-like [Jatropha curcas] |
| 18 |
Hb_000392_210 |
0.2310087783 |
transcription factor |
TF Family: TCP |
hypothetical protein POPTR_0004s04600g [Populus trichocarpa] |
| 19 |
Hb_126933_010 |
0.231752013 |
- |
- |
PREDICTED: reticulon-like protein B5 [Jatropha curcas] |
| 20 |
Hb_004954_020 |
0.2346085828 |
- |
- |
PREDICTED: probable pectinesterase/pectinesterase inhibitor 51 isoform X2 [Jatropha curcas] |