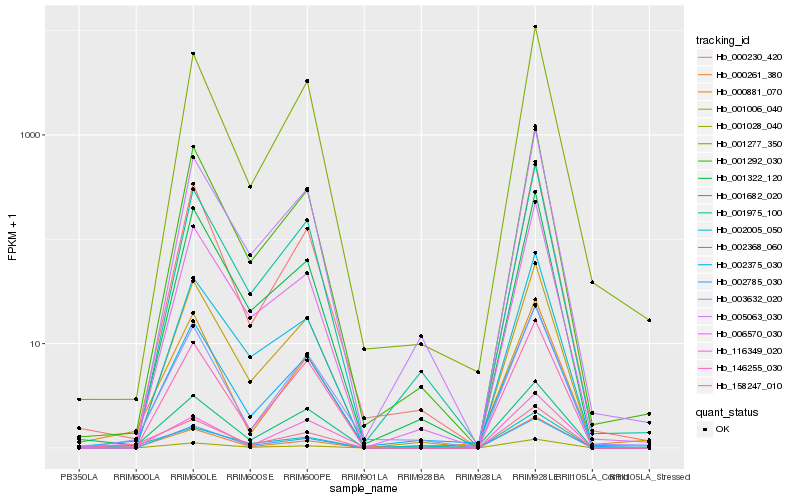
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_158247_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000261_380 |
0.0888079182 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_1019500 [Ricinus communis] |
| 3 |
Hb_002375_030 |
0.1171968558 |
- |
- |
F-box family protein [Populus trichocarpa] |
| 4 |
Hb_002005_050 |
0.1201101297 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 5 |
Hb_001006_040 |
0.1233475323 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g13770, chloroplastic-like isoform X1 [Populus euphratica] |
| 6 |
Hb_000881_070 |
0.1249981232 |
- |
- |
nucleic acid binding protein, putative [Ricinus communis] |
| 7 |
Hb_003632_020 |
0.1256122772 |
- |
- |
calcium-binding family protein [Populus trichocarpa] |
| 8 |
Hb_001292_030 |
0.1256799531 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 9 |
Hb_002785_030 |
0.1257227283 |
- |
- |
Polyketide cyclase/dehydrase and lipid transport superfamily protein [Theobroma cacao] |
| 10 |
Hb_001028_040 |
0.1292288297 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |
| 11 |
Hb_001682_020 |
0.1307843653 |
- |
- |
PREDICTED: uncharacterized protein LOC105628366 [Jatropha curcas] |
| 12 |
Hb_001277_350 |
0.1316332478 |
- |
- |
early light-induced protein, putative [Ricinus communis] |
| 13 |
Hb_001975_100 |
0.1320170629 |
- |
- |
chlorophyll a-b binding protein 151, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000230_420 |
0.1327574255 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 15 |
Hb_006570_030 |
0.13283843 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002368_060 |
0.1338039487 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_116349_020 |
0.1341460849 |
- |
- |
PREDICTED: non-functional NADPH-dependent codeinone reductase 2-like [Jatropha curcas] |
| 18 |
Hb_005063_030 |
0.1347000976 |
- |
- |
Photosystem II reaction center W protein, chloroplast precursor, putative [Ricinus communis] |
| 19 |
Hb_146255_030 |
0.134706105 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HY5-like [Jatropha curcas] |
| 20 |
Hb_001322_120 |
0.1354506884 |
- |
- |
nutrient reservoir, putative [Ricinus communis] |