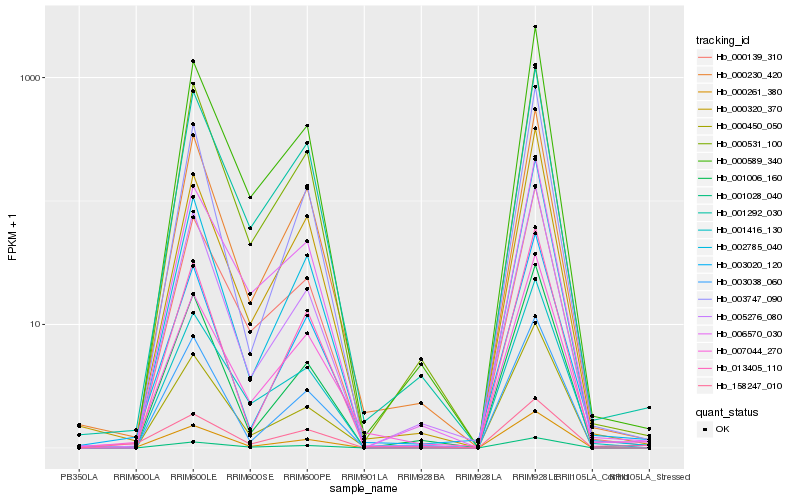
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000261_380 |
0.0 |
transcription factor |
TF Family: ERF |
hypothetical protein RCOM_1019500 [Ricinus communis] |
| 2 |
Hb_000589_340 |
0.0756472662 |
- |
- |
unknown [Populus trichocarpa x Populus deltoides] |
| 3 |
Hb_002785_040 |
0.080047367 |
- |
- |
sedoheptulose-1,7-bisphosphatase, chloroplast, putative [Ricinus communis] |
| 4 |
Hb_000230_420 |
0.0843626827 |
- |
- |
GS [Hevea brasiliensis subsp. brasiliensis] |
| 5 |
Hb_000320_370 |
0.0845825641 |
- |
- |
PREDICTED: ATP synthase gamma chain, chloroplastic [Jatropha curcas] |
| 6 |
Hb_000139_310 |
0.0851886359 |
- |
- |
PREDICTED: glucose-1-phosphate adenylyltransferase large subunit 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000450_050 |
0.0865765845 |
- |
- |
PREDICTED: benzyl alcohol O-benzoyltransferase-like [Jatropha curcas] |
| 8 |
Hb_158247_010 |
0.0888079182 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_001292_030 |
0.0908576721 |
- |
- |
PREDICTED: chlorophyll a-b binding protein CP26, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001416_130 |
0.0915825952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_005276_080 |
0.0915865215 |
- |
- |
PREDICTED: chloroplast stem-loop binding protein of 41 kDa a, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003747_090 |
0.0923613173 |
- |
- |
PREDICTED: glyceraldehyde-3-phosphate dehydrogenase A, chloroplastic isoform X2 [Jatropha curcas] |
| 13 |
Hb_000531_100 |
0.0931697258 |
- |
- |
Photosystem I reaction center subunit III, chloroplast precursor, putative [Ricinus communis] |
| 14 |
Hb_013405_110 |
0.0938279326 |
- |
- |
multidrug resistance pump, putative [Ricinus communis] |
| 15 |
Hb_003020_120 |
0.0938379357 |
- |
- |
PREDICTED: protein TIC 62, chloroplastic [Populus euphratica] |
| 16 |
Hb_006570_030 |
0.0950075959 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 17 |
Hb_007044_270 |
0.0951021854 |
- |
- |
PREDICTED: uncharacterized protein LOC105638707 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001006_160 |
0.0953128546 |
transcription factor |
TF Family: C2H2 |
nucleic acid binding protein, putative [Ricinus communis] |
| 19 |
Hb_003038_060 |
0.0953201591 |
- |
- |
hypothetical protein POPTR_0007s13050g [Populus trichocarpa] |
| 20 |
Hb_001028_040 |
0.0955926164 |
- |
- |
hypothetical protein POPTR_0018s09860g [Populus trichocarpa] |