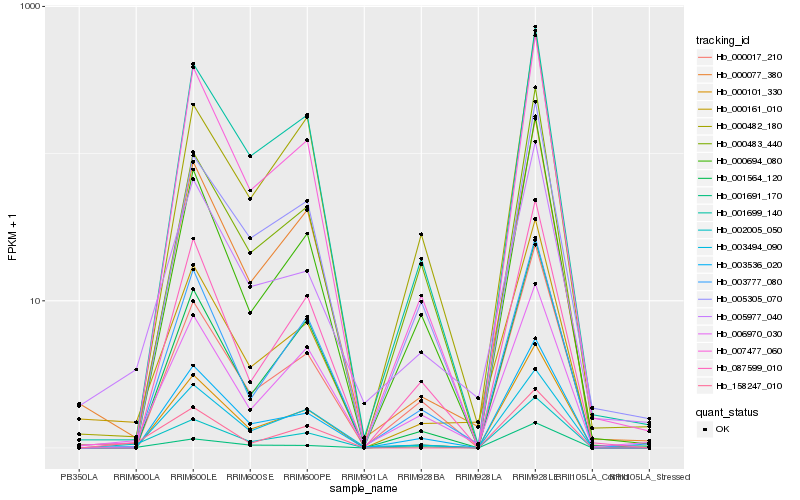
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002005_050 |
0.0 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 2 |
Hb_158247_010 |
0.1201101297 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_006970_030 |
0.1237071575 |
- |
- |
synaptotagmin, putative [Ricinus communis] |
| 4 |
Hb_000161_010 |
0.125753112 |
- |
- |
Ferredoxin-2, chloroplast precursor, putative [Ricinus communis] |
| 5 |
Hb_005305_070 |
0.1283326945 |
- |
- |
hypothetical protein POPTR_0001s26540g [Populus trichocarpa] |
| 6 |
Hb_001699_140 |
0.1293544916 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, leaf isozyme, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000101_330 |
0.1306461275 |
- |
- |
- |
| 8 |
Hb_001691_170 |
0.1331979106 |
- |
- |
PREDICTED: callose synthase 12-like [Glycine max] |
| 9 |
Hb_003494_090 |
0.1337453897 |
transcription factor |
TF Family: MYB |
myb family transcription factor family protein [Populus trichocarpa] |
| 10 |
Hb_003536_020 |
0.1337808147 |
- |
- |
PREDICTED: splicing factor 3A subunit 2 [Jatropha curcas] |
| 11 |
Hb_000483_440 |
0.1352057689 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_000694_080 |
0.1359015279 |
- |
- |
phosphoribulose kinase, putative [Ricinus communis] |
| 13 |
Hb_000017_210 |
0.1365081998 |
- |
- |
PREDICTED: homeobox protein BEL1 homolog [Populus euphratica] |
| 14 |
Hb_087599_010 |
0.1368440118 |
- |
- |
PREDICTED: tropinone reductase homolog [Solanum tuberosum] |
| 15 |
Hb_007477_060 |
0.1373539291 |
- |
- |
unknown [Medicago truncatula] |
| 16 |
Hb_005977_040 |
0.1375125677 |
- |
- |
Photosystem I reaction center subunit IV A [Medicago truncatula] |
| 17 |
Hb_003777_080 |
0.1382707271 |
transcription factor |
TF Family: Trihelix |
PREDICTED: trihelix transcription factor GT-2-like [Populus euphratica] |
| 18 |
Hb_001564_120 |
0.1391162922 |
- |
- |
PREDICTED: peptidyl-prolyl cis-trans isomerase CYP26-2, chloroplastic [Populus euphratica] |
| 19 |
Hb_000482_180 |
0.1402015383 |
- |
- |
PREDICTED: glycine dehydrogenase (decarboxylating), mitochondrial [Jatropha curcas] |
| 20 |
Hb_000077_380 |
0.1405190972 |
- |
- |
PREDICTED: protochlorophyllide reductase [Jatropha curcas] |