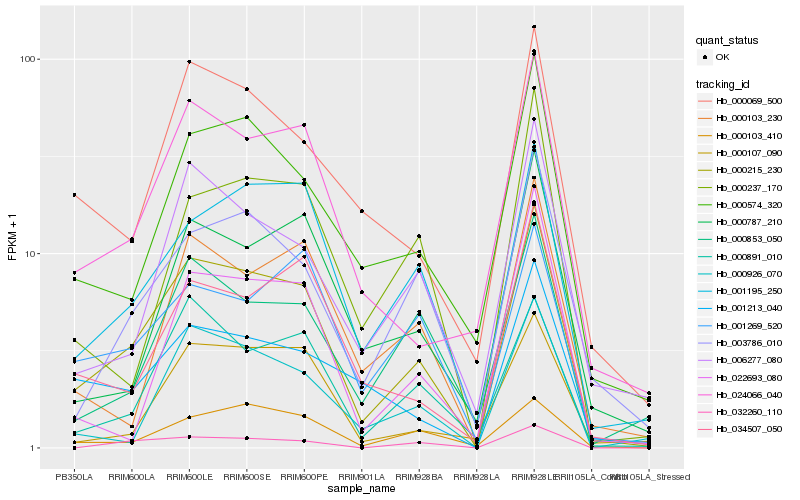
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000215_230 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105637821 [Jatropha curcas] |
| 2 |
Hb_024066_040 |
0.141401893 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 [Jatropha curcas] |
| 3 |
Hb_001195_250 |
0.1440917705 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 4 |
Hb_000574_320 |
0.1511211771 |
- |
- |
hypothetical protein CICLE_v10024937mg [Citrus clementina] |
| 5 |
Hb_003786_010 |
0.1547921566 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 2 isoform X2 [Jatropha curcas] |
| 6 |
Hb_000069_500 |
0.1571014219 |
- |
- |
PREDICTED: rhodanese-like domain-containing protein 4, chloroplastic [Jatropha curcas] |
| 7 |
Hb_000891_010 |
0.1583933382 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_006277_080 |
0.1587290103 |
- |
- |
PREDICTED: uncharacterized protein LOC105649701 [Jatropha curcas] |
| 9 |
Hb_034507_050 |
0.1589233369 |
- |
- |
PREDICTED: uncharacterized protein LOC105646633 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000787_210 |
0.1625751823 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_000103_410 |
0.166155591 |
- |
- |
PREDICTED: putative calcium-transporting ATPase 13, plasma membrane-type [Jatropha curcas] |
| 12 |
Hb_000926_070 |
0.1665600734 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000103_230 |
0.1680188938 |
- |
- |
PREDICTED: chlorophyllase-2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_000107_090 |
0.1702497816 |
- |
- |
PREDICTED: kinesin KP1 [Jatropha curcas] |
| 15 |
Hb_000853_050 |
0.1709935464 |
- |
- |
PREDICTED: putative GTP diphosphokinase RSH1, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_001213_040 |
0.1720496825 |
- |
- |
fructose-1,6-bisphosphatase, putative [Ricinus communis] |
| 17 |
Hb_000237_170 |
0.1723835411 |
transcription factor |
TF Family: HB |
PREDICTED: BEL1-like homeodomain protein 6 [Jatropha curcas] |
| 18 |
Hb_032260_110 |
0.1732931466 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_001269_520 |
0.1753007333 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 20 |
Hb_022693_080 |
0.1764551575 |
- |
- |
PREDICTED: protein NUCLEAR FUSION DEFECTIVE 4 [Jatropha curcas] |