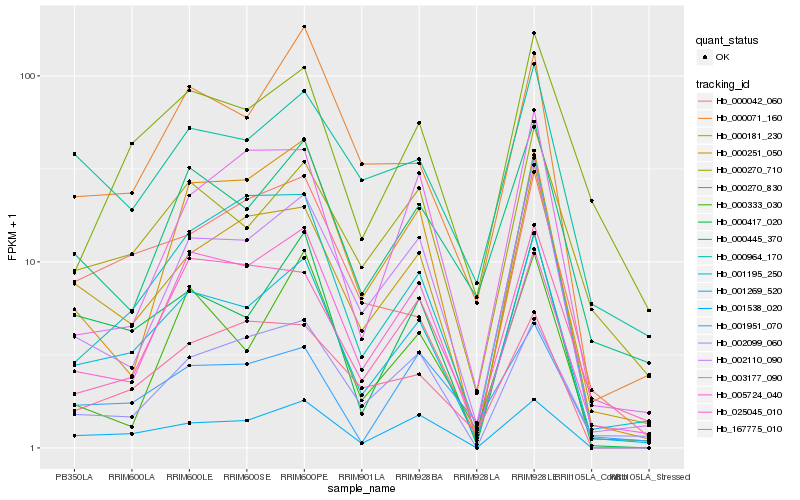
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001269_520 |
0.0 |
- |
- |
PREDICTED: putative receptor-like protein kinase At1g80870 [Jatropha curcas] |
| 2 |
Hb_000251_050 |
0.1110838146 |
- |
- |
hypothetical protein JCGZ_21678 [Jatropha curcas] |
| 3 |
Hb_002110_090 |
0.1163964496 |
- |
- |
PREDICTED: transport inhibitor response 1-like protein [Jatropha curcas] |
| 4 |
Hb_000417_020 |
0.1181809676 |
- |
- |
PREDICTED: uncharacterized protein LOC105649674 [Jatropha curcas] |
| 5 |
Hb_001538_020 |
0.1186419385 |
- |
- |
PREDICTED: uncharacterized protein LOC105629608 [Jatropha curcas] |
| 6 |
Hb_001195_250 |
0.1261897451 |
- |
- |
lrr receptor protein kinase, putative [Ricinus communis] |
| 7 |
Hb_000270_710 |
0.1372483366 |
- |
- |
PREDICTED: WAT1-related protein At5g40240-like isoform X1 [Jatropha curcas] |
| 8 |
Hb_000445_370 |
0.1454390298 |
- |
- |
PREDICTED: probable methyltransferase PMT24 [Jatropha curcas] |
| 9 |
Hb_000071_160 |
0.1483553358 |
- |
- |
PREDICTED: protein IQ-DOMAIN 14 [Jatropha curcas] |
| 10 |
Hb_002099_060 |
0.150978448 |
- |
- |
Protein AFR, putative [Ricinus communis] |
| 11 |
Hb_000270_830 |
0.1515697037 |
- |
- |
aldehyde dehydrogenase, putative [Ricinus communis] |
| 12 |
Hb_000964_170 |
0.1541472112 |
- |
- |
Cellulose synthase A catalytic subunit 6 [UDP-forming], putative [Ricinus communis] |
| 13 |
Hb_005724_040 |
0.1542983394 |
- |
- |
hypothetical protein JCGZ_13142 [Jatropha curcas] |
| 14 |
Hb_025045_010 |
0.1577473292 |
- |
- |
catalytic, putative [Ricinus communis] |
| 15 |
Hb_000181_230 |
0.1593553685 |
- |
- |
copper ion binding protein, putative [Ricinus communis] |
| 16 |
Hb_000042_060 |
0.1611580926 |
- |
- |
PREDICTED: uncharacterized protein LOC105632794 [Jatropha curcas] |
| 17 |
Hb_167775_010 |
0.163665366 |
- |
- |
PREDICTED: putative disease resistance protein RGA3 [Jatropha curcas] |
| 18 |
Hb_000333_030 |
0.1650618412 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 19 |
Hb_003177_090 |
0.1653724785 |
- |
- |
PREDICTED: filament-like plant protein 4 [Jatropha curcas] |
| 20 |
Hb_001951_070 |
0.1661845971 |
- |
- |
putative protein [Arabidopsis thaliana] |